7KED
 
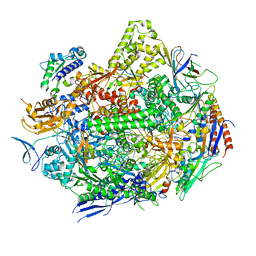 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KEE
 
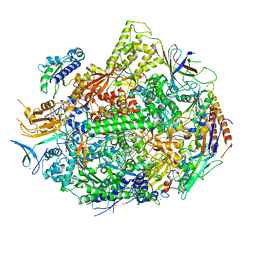 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | Descriptor: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
6SNJ
 
 | |
3BY4
 
 | |
6T6K
 
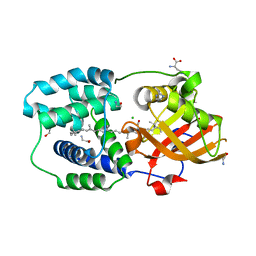 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCINE, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T6M
 
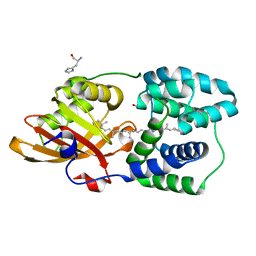 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | Descriptor: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
3C0R
 
 | |
2N4P
 
 | | Solution structure of the n-terminal domain of tdp-43 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F, Buratti, E, Laurents, D.V. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The TDP-43 N-terminal domain structure at high resolution.
Febs J., 283, 2016
|
|
8DMB
 
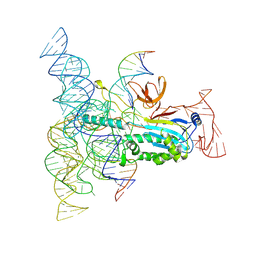 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
8EL8
 
 | |
8EL7
 
 | |
3P23
 
 | |
3RTV
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with natural primer/template DNA | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Marx, A, Diederichs, K, Betz, K. | | Deposit date: | 2011-05-04 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3S04
 
 | |
6ZJF
 
 | | Crystal structure of STK17B (DRAK2) in complex with AP-229 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-cyclopropylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Picado, A, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6ZDW
 
 | |
7AKG
 
 | |
8UL9
 
 | | Cholinephosphotransferase in complex with diacylglycerol | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cholinephosphotransferase 1, DIACYL GLYCEROL, ... | | Authors: | Roberts, J.R, Maeda, S, Ohi, M.D. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for catalysis and selectivity of phospholipid synthesis by eukaryotic choline-phosphotransferase.
Nat Commun, 16, 2025
|
|
8OXR
 
 | |
8PWW
 
 | | PfRH5 bound to monoclonal antibody MAD8-151 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Farrell, B, Higgins, M.K. | | Deposit date: | 2023-07-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8PWU
 
 | | PfRH5 bound to monoclonal antibody MAD10-255 | | Descriptor: | Reticulocyte-binding protein homolog 5, monoclonal antibody MAD10-255 | | Authors: | Farrell, B, Higgins, M.K. | | Deposit date: | 2023-07-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8Q5D
 
 | | PfRH5 bound to monoclonal antibody MAD10-466 | | Descriptor: | Monoclonal antibody MAD10-466 heavy chain, Monoclonal antibody MAD10-466 light chain, Reticulocyte-binding protein homolog 5 | | Authors: | Farrell, B, Higgins, M.K. | | Deposit date: | 2023-08-08 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8PWV
 
 | | PfRH5 bound to monoclonal antibody MAD8-502 | | Descriptor: | Reticulocyte-binding protein homolog 5, monoclonal antibody MAD8-502 | | Authors: | Farrell, B, Higgins, M.K. | | Deposit date: | 2023-07-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8PWX
 
 | | PfRH5 bound to monoclonal antibody R5.008 | | Descriptor: | GLYCEROL, Reticulocyte-binding protein homolog 5, scFv fragment for antibody R5.008 | | Authors: | Farrell, B, Higgins, M.K. | | Deposit date: | 2023-07-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
7SF8
 
 | | GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer | | Descriptor: | G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Barros-Alvarez, X, Panova, O, Skiniotis, G. | | Deposit date: | 2021-10-03 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The tethered peptide activation mechanism of adhesion GPCRs.
Nature, 604, 2022
|
|
