2QUM
 
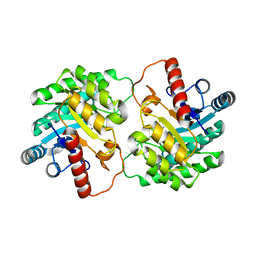 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii with D-tagatose | | Descriptor: | D-tagatose, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2FAU
 
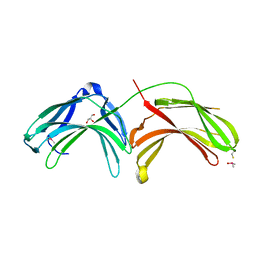 | | Crystal structure of human vps26 | | Descriptor: | GLYCEROL, Vacuolar protein sorting 26 | | Authors: | Shi, H, Rojas, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7VSG
 
 | |
7VSH
 
 | |
2MQE
 
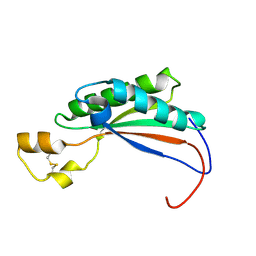 | |
1UF8
 
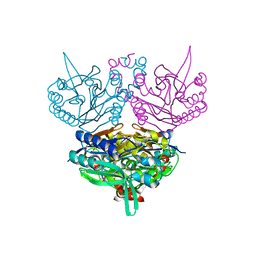 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-Phenylalanine | | Descriptor: | D-[(AMINO)CARBONYL]PHENYLALANINE, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF4
 
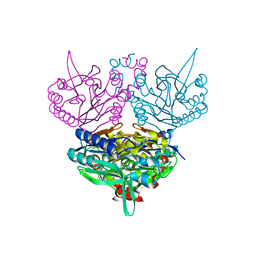 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase | | Descriptor: | N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid
To be published
|
|
1UF7
 
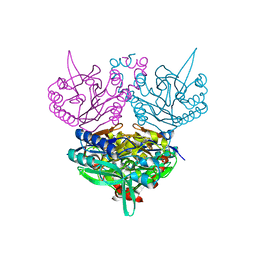 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-valine | | Descriptor: | 3-METHYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF5
 
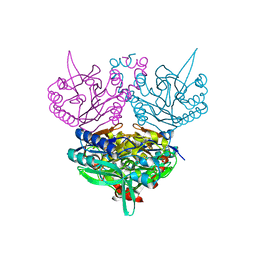 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
3VKM
 
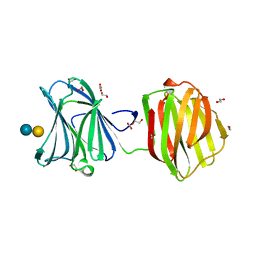 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
5XGC
 
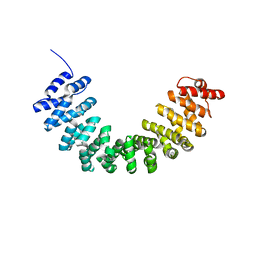 | | Crystal structure of SmgGDS-558 | | Descriptor: | Rap1 GTPase-GDP dissociation stimulator 1 | | Authors: | Shimizu, H, Toma-Fukai, S, Shimizu, T. | | Deposit date: | 2017-04-13 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based analysis of the guanine nucleotide exchange factor SmgGDS reveals armadillo-repeat motifs and key regions for activity and GTPase binding
J. Biol. Chem., 292, 2017
|
|
2OHD
 
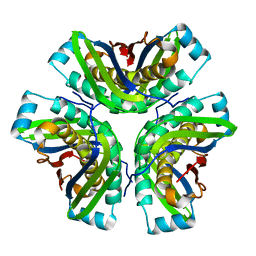 | | Crystal structure of hypothetical molybdenum cofactor biosynthesis protein C from Sulfolobus tokodaii | | Descriptor: | Probable molybdenum cofactor biosynthesis protein C | | Authors: | Yoshida, H, Yamada, M, Kuramitsu, S, Kamitori, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative molybdenum-cofactor biosynthesis protein C (MoaC) from Sulfolobus tokodaii (ST0472)
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1UMG
 
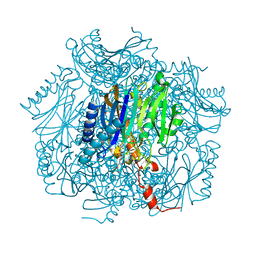 | | Crystal structure of fructose-1,6-bisphosphatase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), 385aa long conserved hypothetical protein, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-09-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Structure, 12, 2004
|
|
5XH7
 
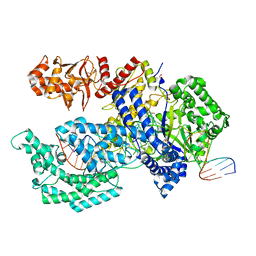 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RR variant in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XH6
 
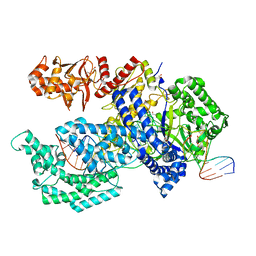 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
1RFP
 
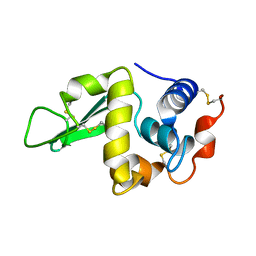 | |
5ZFS
 
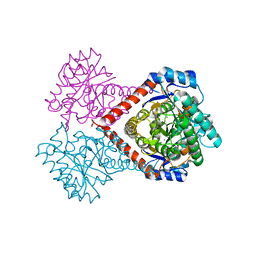 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | Descriptor: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5ZHX
 
 | |
3VSK
 
 | |
2Z5C
 
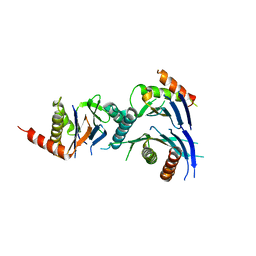 | | Crystal Structure of a Novel Chaperone Complex for Yeast 20S Proteasome Assembly | | Descriptor: | Proteasome component PUP2, Protein YPL144W, Uncharacterized protein YLR021W | | Authors: | Yashiroda, H, Mizushima, T, Okamoto, K, Kameyama, T, Hayashi, H, Kishimoto, T, Kasahara, M, Kurimoto, E, Sakata, E, Suzuki, A, Hirano, Y, Murata, S, Kato, K, Yamane, T, Tanaka, K. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
3VSL
 
 | | Crystal structure of penicillin-binding protein 3 (PBP3) from methicilin-resistant Staphylococcus aureus in the cefotaxime bound form. | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Yoshida, H, Tame, J.R, Park, S.Y. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-Binding Protein 3 (PBP3) from Methicillin-Resistant Staphylococcus aureus in the Apo and Cefotaxime-Bound Forms.
J.Mol.Biol., 423, 2012
|
|
3WXR
 
 | | Yeast 20S proteasome with a mutation of alpha7 subunit | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Yashiroda, H, Toda, Y, Otsu, S, Takagi, K, Mizushima, T, Murata, S. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | N-terminal alpha 7 deletion of the proteasome 20S core particle substitutes for yeast PI31 function
Mol.Cell.Biol., 35, 2015
|
|
3WV6
 
 | |
2Z5B
 
 | | Crystal Structure of a Novel Chaperone Complex for Yeast 20S Proteasome Assembly | | Descriptor: | Protein YPL144W, Uncharacterized protein YLR021W | | Authors: | Yashiroda, H, Mizushima, T, Okamoto, K, Kameyama, T, Hayashi, H, Kishimoto, T, Kasahara, M, Kurimoto, E, Sakata, E, Suzuki, A, Hirano, Y, Murata, S, Kato, K, Yamane, T, Tanaka, K. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
