2RQR
 
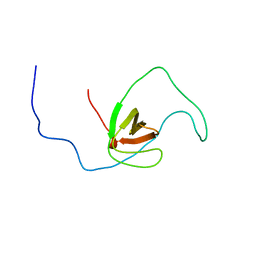 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | 分子名称: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | 著者 | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-10-21 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2DVW
 
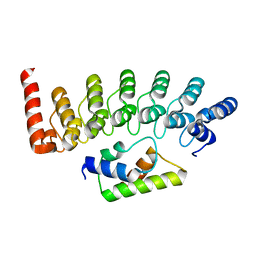 | |
2DZN
 
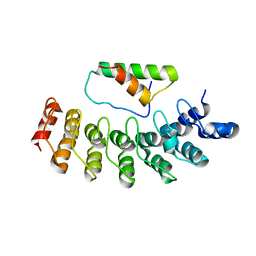 | |
1N27
 
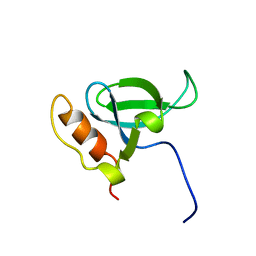 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | 分子名称: | Hepatoma-derived growth factor, related protein 3 | | 著者 | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-10-22 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
3BOY
 
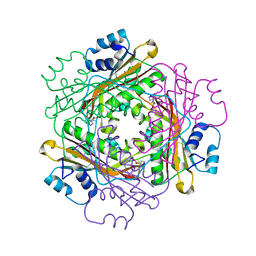 | | Crystal structure of the HutP antitermination complex bound to the HUT mRNA | | 分子名称: | 5'-R(*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | 著者 | Kumarevel, T.S, Balasundaresan, D, Jeyakanthan, J, Shinkai, A, Yokoyama, S, Kumar, P.K.R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-12-18 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of HutP complexed with the 55-mer RNA
To be Published
|
|
1FNX
 
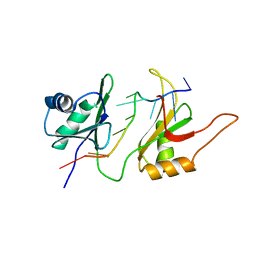 | | SOLUTION STRUCTURE OF THE HUC RBD1-RBD2 COMPLEXED WITH THE AU-RICH ELEMENT | | 分子名称: | AU-RICH RNA ELEMENT, HU ANTIGEN C | | 著者 | Inoue, M, Hirao, M, Kasashima, K, Kim, I.-S, Kawai, G, Kigawa, T, Sakamoto, H, Muto, Y, Yokoyama, S. | | 登録日 | 2000-08-24 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of mouse HuC RNA-binding domains complexed with an AU-Rich element reveals determinants of neuronal differentiation
To be Published, 2000
|
|
2GGF
 
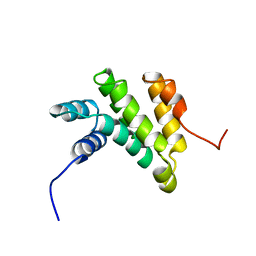 | | Solution structure of the MA3 domain of human Programmed cell death 4 | | 分子名称: | Programmed cell death 4, isoform 1 | | 著者 | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-24 | | 公開日 | 2007-04-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the MA3 domain of human Programmed cell death 4
To be Published
|
|
3UG0
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the simplified genetic code | | 分子名称: | Green fluorescent protein | | 著者 | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | 登録日 | 2011-11-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
2QQ3
 
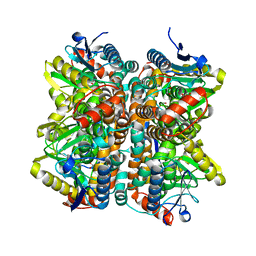 | | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426 | | 分子名称: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase subunit I | | 著者 | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-07-26 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426
To be Published
|
|
6LR2
 
 | |
5FWM
 
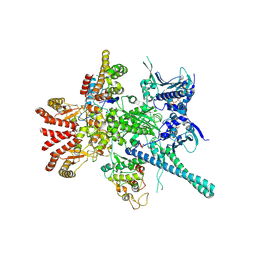 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | 著者 | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | 登録日 | 2016-02-18 | | 公開日 | 2016-07-06 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | 著者 | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | 登録日 | 2017-02-20 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
5FWK
 
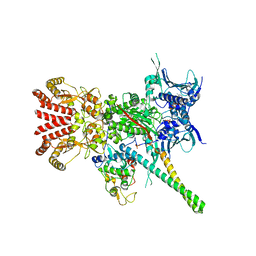 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | 著者 | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | 登録日 | 2016-02-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2019-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWP
 
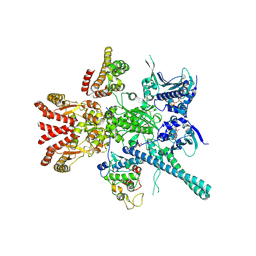 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | 著者 | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | 登録日 | 2016-02-18 | | 公開日 | 2016-10-26 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Atomic Structure of Hsp90:Cdc37:Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWL
 
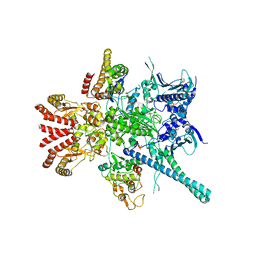 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | 著者 | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | 登録日 | 2016-02-18 | | 公開日 | 2016-07-06 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
2R1G
 
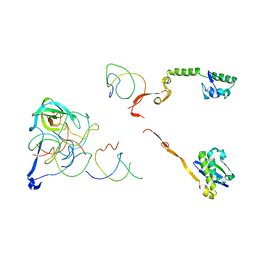 | | Coordinates of the thermus thermophilus 30S components neighboring RbfA as obtained by fitting into the CRYO-EM map of A 30S-RBFA complex | | 分子名称: | 16S RIBOSOMAL RNA HELIX 1, 16S RIBOSOMAL RNA HELIX 18, 16S RIBOSOMAL RNA HELIX 27, ... | | 著者 | Datta, P.P, Wilson, D.N, Kawazoe, M, Swami, N.K, Kaminishi, T, Sharma, M.R, Booth, T.M, Takemoto, C, Fucini, P, Yokoyama, S, Agrawal, R.K. | | 登録日 | 2007-08-22 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (12.5 Å) | | 主引用文献 | Structural aspects of RbfA action during small ribosomal subunit assembly.
Mol.Cell, 28, 2007
|
|
2R1C
 
 | | Coordinates of the thermus thermophilus ribosome binding factor A (RbfA) homology model as fitted into the CRYO-EM map of a 30S-RBFA complex | | 分子名称: | Ribosome-binding factor A | | 著者 | Datta, P.P, Wilson, D.N, Kawazoe, M, Swami, N.K, Kaminishi, T, Sharma, M.R, Booth, T.M, Takemoto, C, Fucini, P, Yokoyama, S, Agrawal, R.K. | | 登録日 | 2007-08-22 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (12.5 Å) | | 主引用文献 | Structural aspects of RbfA action during small ribosomal subunit assembly.
Mol.Cell, 28, 2007
|
|
5X50
 
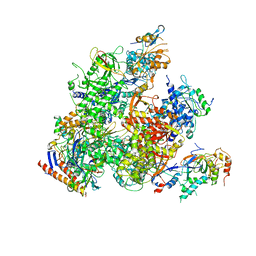 | | RNA Polymerase II from Komagataella Pastoris (Type-2 crystal) | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | 著者 | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.293 Å) | | 主引用文献 | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5X51
 
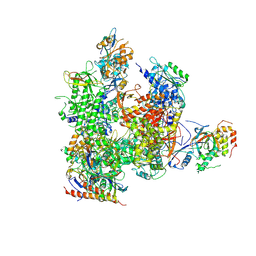 | | RNA Polymerase II from Komagataella Pastoris (Type-3 crystal) | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | 著者 | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-17 | | 実験手法 | X-RAY DIFFRACTION (6.996 Å) | | 主引用文献 | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5XAX
 
 | |
4WRI
 
 | | Crystal structure of okadaic acid binding protein 2.1 | | 分子名称: | OKADAIC ACID, Okadaic acid binding protein 2-alpha | | 著者 | Ehara, H, Makino, M, Kodama, K, Ito, T, Sekine, S, Fukuzawa, S, Yokoyama, S, Tachibana, K. | | 登録日 | 2014-10-24 | | 公開日 | 2015-05-27 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Okadaic Acid Binding Protein 2.1: A Sponge Protein Implicated in Cytotoxin Accumulation
Chembiochem, 16, 2015
|
|
5WRK
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y608 peptide | | 分子名称: | AP-2 complex subunit mu, Insulin receptor substrate 1, NICKEL (II) ION | | 著者 | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | 登録日 | 2016-12-02 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
4WR5
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | 分子名称: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | 著者 | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | 登録日 | 2014-10-23 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WR4
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | 分子名称: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | 著者 | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | 登録日 | 2014-10-23 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WQS
 
 | | Thermus thermophilus RNA polymerase backtracked complex | | 分子名称: | DNA (28-MER), DNA (5'-D(P*GP*TP*AP*GP*CP*TP*TP*GP*TP*GP*GP*TP*AP*GP*TP*GP*AP*CP*GP*AP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Murayama, Y, Sekine, S, Yokoyama, S. | | 登録日 | 2014-10-22 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4.306 Å) | | 主引用文献 | The Ratcheted and Ratchetable Structural States of RNA Polymerase Underlie Multiple Transcriptional Functions.
Mol.Cell, 57, 2015
|
|
