5YXJ
 
 | | FXR ligand binding domain | | 分子名称: | 2-[benzyl(methyl)amino]ethyl methyl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | 著者 | Yi, L, Yong, L. | | 登録日 | 2017-12-05 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | A ligand of drug binding to FXR
To Be Published
|
|
5YXB
 
 | | A ligand binding to FXR | | 分子名称: | 2-methoxyethyl (2E)-3-phenylprop-2-en-1-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | 著者 | Yi, L, Yong, L. | | 登録日 | 2017-12-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | A ligand binding to FXR
To Be Published
|
|
5YXL
 
 | | A ligand M binding to FXR | | 分子名称: | 2-methoxyethyl propan-2-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | 著者 | Yi, L, Yong, L. | | 登録日 | 2017-12-05 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | A ligand M binding to FXR
To Be Published
|
|
5YXD
 
 | | A ligand F binding to FXR | | 分子名称: | Bile acid receptor, Peptide from Nuclear receptor coactivator, ethyl methyl 4-(2,3-dichlorophenyl)-2,6-dimethylpyridine-3,5-dicarboxylate | | 著者 | Yi, L, Yong, L. | | 登録日 | 2017-12-05 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | A ligand F binding to FXR
To Be Published
|
|
5YXU
 
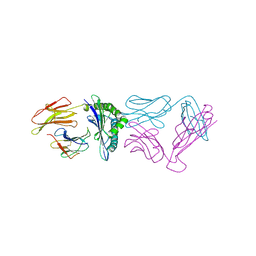 | |
3ELO
 
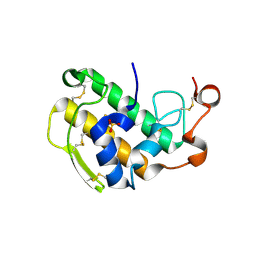 | | Crystal Structure of Human Pancreatic Prophospholipase A2 | | 分子名称: | Phospholipase A2, SULFATE ION | | 著者 | Xu, W, Yi, L, Feng, Y, Chen, L, Liu, J. | | 登録日 | 2008-09-22 | | 公開日 | 2009-04-14 | | 最終更新日 | 2014-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural insight into the activation mechanism of human pancreatic prophospholipase A2
J.Biol.Chem., 284, 2009
|
|
5SYD
 
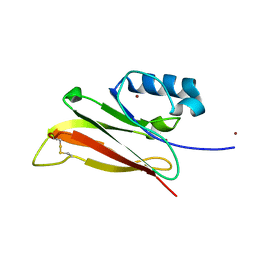 | |
4LCT
 
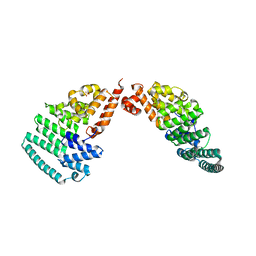 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | 分子名称: | COP9 signalosome complex subunit 1, SULFATE ION | | 著者 | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | 登録日 | 2013-06-23 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5ZG9
 
 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | 著者 | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | 登録日 | 2018-03-08 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
6JBI
 
 | | Structure of Tps1 apo structure | | 分子名称: | Trehalose-6-phosphate synthase | | 著者 | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | 登録日 | 2019-01-25 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
5C5P
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-3-(1-hydroxy-2-methylpropan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5R
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (7R)-2-hydroxy-7-(propan-2-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine-3-carbonitrile, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-10-methyl-3-(propan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
2R8Z
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in complex with a phosphate and a calcium ion | | 分子名称: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | 登録日 | 2007-09-11 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
2RGZ
 
 | | Ensemble refinement of the protein crystal structure of human heme oxygenase-2 C127A (HO-2) with bound heme | | 分子名称: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2007-10-05 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
2R8X
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli | | 分子名称: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | 登録日 | 2007-09-11 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
2R8Y
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in a complex with Ca | | 分子名称: | CALCIUM ION, CHLORIDE ION, YrbI from Escherichia coli | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | 登録日 | 2007-09-11 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
2R8E
 
 | | Crystal structure of YrbI from Escherichia coli in complex with Mg | | 分子名称: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION, MAGNESIUM ION | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R, Biswas, T. | | 登録日 | 2007-09-10 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
3I6B
 
 | |
3HYC
 
 | |
2P5W
 
 | | Crystal structures of high affinity human T-cell receptors bound to pMHC reveal native diagonal binding geometry | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, CALCIUM ION, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-03-16 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
2PYF
 
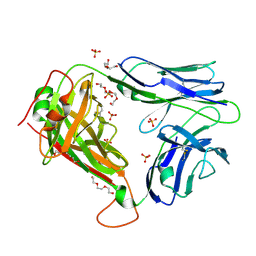 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry Unbound TCR Clone 5-1 | | 分子名称: | SULFATE ION, T-Cell Receptor, Alpha Chain, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-05-16 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
2P5E
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC Reveal Native Diagonal Binding Geometry | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-03-15 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
2Q32
 
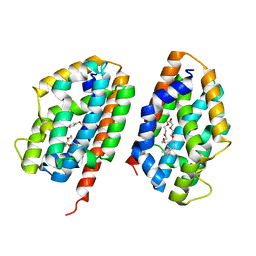 | | Crystal structure of human heme oxygenase-2 C127A (HO-2) | | 分子名称: | Heme oxygenase 2, OXTOXYNOL-10 | | 著者 | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2007-05-29 | | 公開日 | 2007-06-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
2PYE
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry TCR Clone C5C1 Complexed with MHC | | 分子名称: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-05-16 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
