6TH6
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | 登録日 | 2019-11-18 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
6SKF
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | 登録日 | 2019-08-15 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
6SKG
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome in TkNat10 deleted strain | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | 登録日 | 2019-08-15 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
8RNZ
 
 | |
4N5B
 
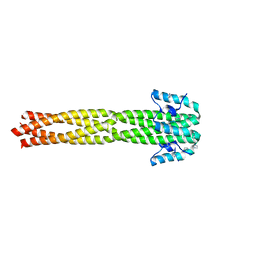 | | Crystal structure of the Nipah virus phosphoprotein tetramerization domain | | 分子名称: | IMIDAZOLE, Phosphoprotein | | 著者 | Bruhn, J.F, Barnett, K, Bibby, J, Thomas, J, Keegan, R, Rigden, D, Bornholdt, Z.A, Saphire, E.O. | | 登録日 | 2013-10-09 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the nipah virus phosphoprotein tetramerization domain.
J.Virol., 88, 2014
|
|
5HP2
 
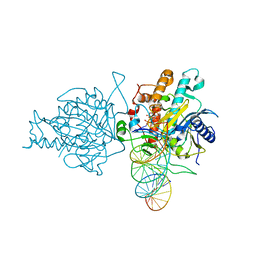 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AU basepair at reaction site | | 分子名称: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*UP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | 著者 | Matthews, M.M, Fisher, A.J, Beal, P.A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.983 Å) | | 主引用文献 | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HP3
 
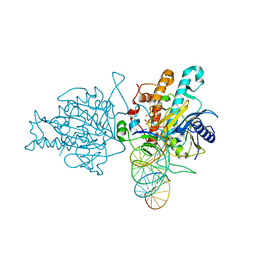 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AC mismatch at reaction site | | 分子名称: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | 著者 | Matthews, M.M, Fisher, A.J, Beal, P.A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.091 Å) | | 主引用文献 | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5ED2
 
 | |
5ED1
 
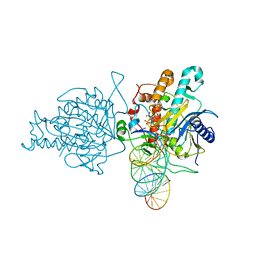 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from S. cerevisiae BDF2 gene | | 分子名称: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | 著者 | Matthews, M.M, Fisher, A.J, Beal, P.A. | | 登録日 | 2015-10-20 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6VFF
 
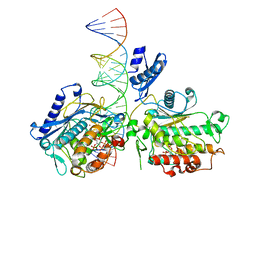 | | Dimer of Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from human GLI1 gene | | 分子名称: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*UP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | 著者 | Thuy-boun, A.S, Fisher, A.J, Beal, P.A. | | 登録日 | 2020-01-03 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition.
Nucleic Acids Res., 48, 2020
|
|
6D06
 
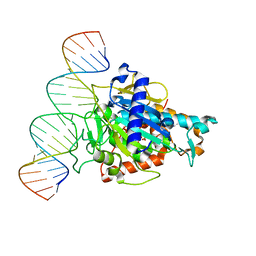 | | Human ADAR2d E488Y mutant complexed with dsRNA containing an abasic site opposite the edited base | | 分子名称: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*CP*AP*GP*AP*GP*CP*CP*CP*CP*CP*NP*AP*GP*CP*AP*UP*CP*GP*CP*GP*AP*GP*C)-3'), ... | | 著者 | Matthews, M.M, Fisher, A.J, Beal, P.A. | | 登録日 | 2018-04-10 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | A Bump-Hole Approach for Directed RNA Editing.
Cell Chem Biol, 26, 2019
|
|
6B0D
 
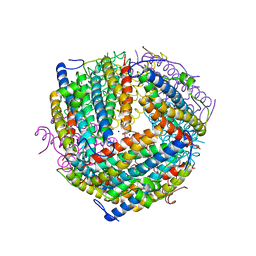 | | An E. coli DPS protein from ferritin superfamily | | 分子名称: | DNA protection during starvation protein, FORMIC ACID, SODIUM ION | | 著者 | Rui, W, Ruslan, S, Ronan, K, Adam, J.S. | | 登録日 | 2017-09-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6M
 
 | | Cyanase from Serratia proteamaculans | | 分子名称: | Cyanate hydratase | | 著者 | Xu, Y. | | 登録日 | 2017-10-02 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BY0
 
 | |
