4BXO
 
 | | Architecture and DNA recognition elements of the Fanconi anemia FANCM- FAAP24 complex | | 分子名称: | 5'-D(*GP*AP*TP*GP*AP*TP*GP*CP*TP*GP*CP)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*AP*TP*CP)-3', CALCIUM ION, ... | | 著者 | Coulthard, R, Deans, A, Swuec, P, Bowles, M, Purkiss, A, Costa, A, West, S, McDonald, N. | | 登録日 | 2013-07-15 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Architecture and DNA Recognition Elements of the Fanconi Anemia Fancm-Faap24 Complex.
Structure, 21, 2013
|
|
6HUD
 
 | |
6S6S
 
 | |
6S6X
 
 | | Structure of Azospirillum brasilense Glutamate Synthase in a6b6 oligomeric state. | | 分子名称: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Chaves-Sanjuan, A, Camilloni, C, Bolognesi, M. | | 登録日 | 2019-07-03 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Structures of Azospirillum brasilense Glutamate Synthase in Its Oligomeric Assemblies.
J.Mol.Biol., 431, 2019
|
|
6S6T
 
 | |
6S6U
 
 | |
7ZPP
 
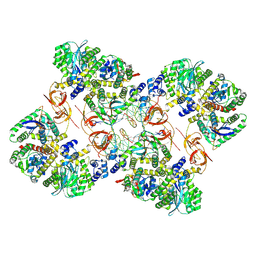 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | 分子名称: | Integrase, vDNA, non-transferred strand, ... | | 著者 | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | 登録日 | 2022-04-28 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
8R7H
 
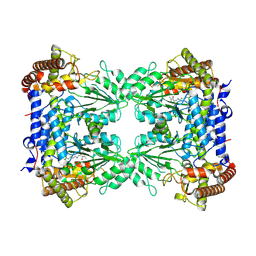 | | Cryo-EM structure of Human SHMT1 | | 分子名称: | Serine hydroxymethyltransferase, cytosolic | | 著者 | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | 登録日 | 2023-11-24 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
6YP7
 
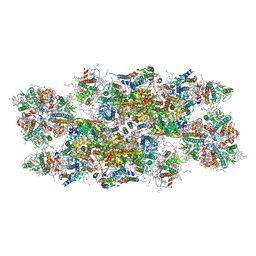 | | PSII-LHCII C2S2 supercomplex from Pisum sativum grown in high light conditions | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Grinzato, A, Albanese, P, Zanotti, G, Pagliano, C. | | 登録日 | 2020-04-15 | | 公開日 | 2020-11-25 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | High-Light versus Low-Light: Effects on Paired Photosystem II Supercomplex Structural Rearrangement in Pea Plants.
Int J Mol Sci, 21, 2020
|
|
5T3A
 
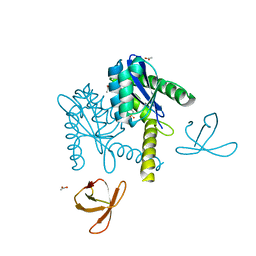 | |
4U7D
 
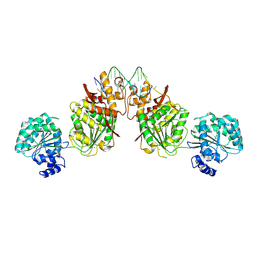 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | 分子名称: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | 著者 | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | 登録日 | 2014-07-30 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5M0R
 
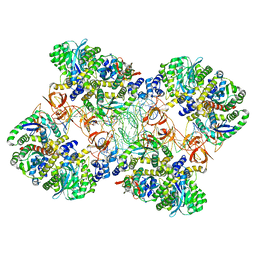 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | 分子名称: | integrase, tDNA, vDNA, ... | | 著者 | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | 登録日 | 2016-10-05 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
5LLJ
 
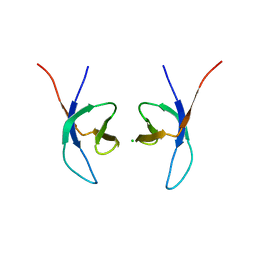 | |
8A11
 
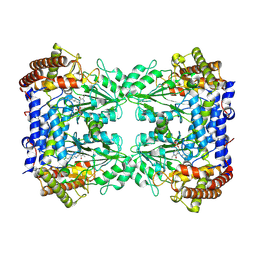 | | Cryo-EM structure of the Human SHMT1-RNA complex | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | 著者 | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | 登録日 | 2022-05-30 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
7ZTH
 
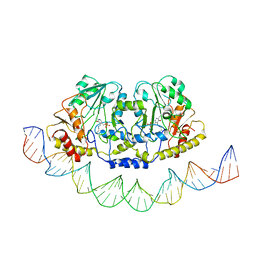 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-05-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
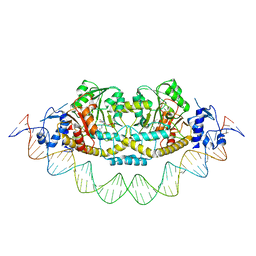 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-20 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
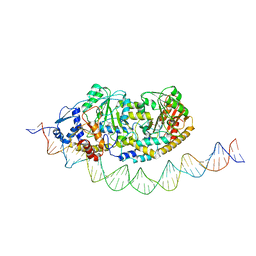 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | 登録日 | 2022-04-14 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
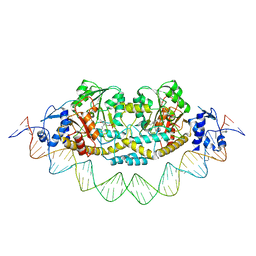 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-27 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7Q54
 
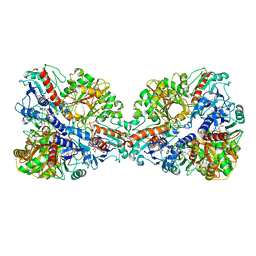 | | Single Particle Cryo-EM structure of photosynthetic A4B4-glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia. | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | 著者 | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q55
 
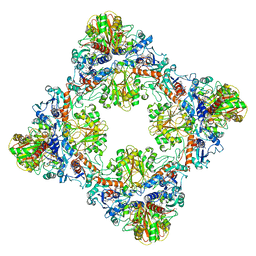 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase hexadecamer (major conformer) from Spinacia oleracia. | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | 著者 | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q56
 
 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase (minor conformer) from Spinacia oleracea. | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | 著者 | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q57
 
 | | Single Particle Cryo-EM structure of photosynthetic A10B10 glyceraldehyde-3-phospahte dehydrogenase from Spinacia oleracea. | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | 著者 | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (13 Å) | | 主引用文献 | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q53
 
 | | Single Particle Cryo-EM structure of photosynthetic A2B2 glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | 著者 | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7NP4
 
 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
