8ZJD
 
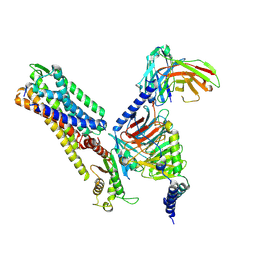 | | Cryo-EM structure of kisspeptin receptor bound to KP-10 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(q) subunit alpha, ... | | 著者 | Shen, S, Liu, H, Xu, H.E. | | 登録日 | 2024-05-14 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Cryo-EM structure of kisspeptin receptor bound to KP-10
To Be Published
|
|
8ZJE
 
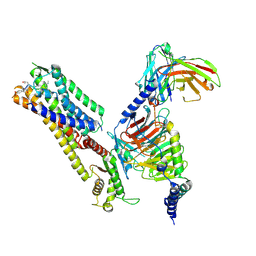 | | Cryo-EM structure of kisspeptin receptor bound to TAK-448 | | 分子名称: | ACY-DTY-HYP-ASN-THR-PHE-XZA-LEU-NMM-TRP-NH2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Shen, S, Liu, H, Xu, H.E. | | 登録日 | 2024-05-14 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Cryo-EM structure of kisspeptin receptor bound to TAK-448
To Be Published
|
|
7LK0
 
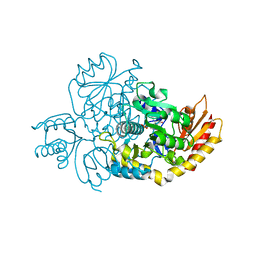 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | 分子名称: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | 著者 | Butrin, A, Shen, S, Liu, D, Silverman, R. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LK1
 
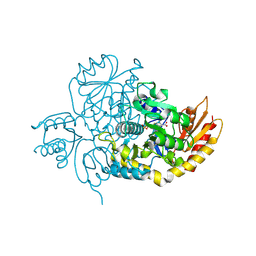 | | Ornithine Aminotransferase (OAT) with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) - 1 Hour Soaking | | 分子名称: | (1R,4R)-4-fluoro-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-2-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | 著者 | Butrin, A, Shen, S, Liu, D, Silverman, R. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
8EZ1
 
 | | Human Ornithine Aminotransferase (hOAT) co-crystallized with its inactivator 3-Amino-4-fluorocyclopentenecarboxylic Acid | | 分子名称: | (1R,3S,4Z)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, (3E,4E)-4-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-iminocyclopent-1-ene-1-carboxylic acid, Ornithine aminotransferase, ... | | 著者 | Butrin, A, Shen, S, Silverman, R, Liu, D. | | 登録日 | 2022-10-30 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural and Mechanistic Basis for the Inactivation of Human Ornithine Aminotransferase by (3 S ,4 S )-3-Amino-4-fluorocyclopentenecarboxylic Acid.
Molecules, 28, 2023
|
|
6V8C
 
 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | 分子名称: | 3-aminocyclohexa-1,3-diene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | 著者 | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | 登録日 | 2019-12-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
6V8D
 
 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | 分子名称: | (3Z)-3-iminocyclohex-1-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | 著者 | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | 登録日 | 2019-12-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
6AJK
 
 | | Crystal structure of TFB1M and h45 in homo sapiens | | 分子名称: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | 著者 | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | 登録日 | 2018-08-28 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AAX
 
 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | 分子名称: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | 著者 | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | 登録日 | 2018-07-19 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.994 Å) | | 主引用文献 | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
7JX9
 
 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | 分子名称: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | 著者 | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | 登録日 | 2020-08-26 | | 公開日 | 2021-01-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
7CNC
 
 | | cystal structure of human ERH in complex with DGCR8 | | 分子名称: | Enhancer of rudimentary homolog, Microprocessor complex subunit DGCR8 | | 著者 | Li, F, Shen, S. | | 登録日 | 2020-07-30 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8.
Nucleic Acids Res., 48, 2020
|
|
7T1Q
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid | | 分子名称: | ACETATE ION, SUCCINIC ACID, Succinyl-diaminopimelate desuccinylase, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-02 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid.
To be Published
|
|
5VSZ
 
 | |
5VSX
 
 | |
6NAU
 
 | | 1.55 Angstrom Resolution Crystal Structure of 6-phosphogluconolactonase from Klebsiella pneumoniae | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-phosphogluconolactonase, CHLORIDE ION | | 著者 | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-12-06 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DT3
 
 | | 1.2 Angstrom Resolution Crystal Structure of Nucleoside Triphosphatase NudI from Klebsiella pneumoniae in Complex with HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoside triphosphatase NudI | | 著者 | Minasov, G, Shuvalova, L, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-15 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7KP2
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | 分子名称: | L-NEOPTERIN, Putative Pterin Binding Protein | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-10 | | 公開日 | 2021-11-17 | | 最終更新日 | 2022-08-17 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
7KOS
 
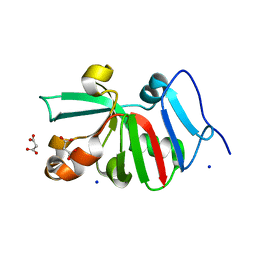 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | 分子名称: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-09 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7KOU
 
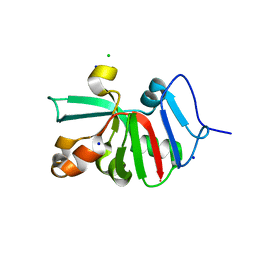 | | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pterin Binding Protein, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-10 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7RJ3
 
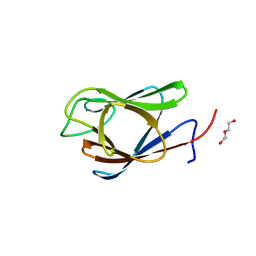 | | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis | | 分子名称: | DI(HYDROXYETHYL)ETHER, Glycogen accumulation regulator GarA | | 著者 | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis.
To Be Published
|
|
5I4C
 
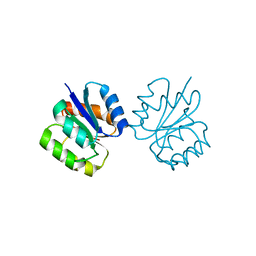 | | Crystal structure of non-phosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli | | 分子名称: | Transcriptional regulatory protein RcsB | | 著者 | Filippova, E.V, Wawrzak, Z, Minasov, G, Ruan, J, Pshenychnyi, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-11 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli.
Protein Sci., 25, 2016
|
|
8F8O
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-11-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids
To Be Published
|
|
6MUK
 
 | | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae. | | 分子名称: | Peptidase M23, ZINC ION | | 著者 | Minasov, G, Shuvalova, L, Pshenychnyi, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.
To Be Published
|
|
6R0K
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with a inhibitor SS208 | | 分子名称: | 5-[2-[(3,4-dichlorophenyl)carbonylamino]ethyl]-~{N}-oxidanyl-1,2-oxazole-3-carboxamide, Histone deacetylase 6, POTASSIUM ION, ... | | 著者 | Barinka, C, Ustinova, K, Motlova, L, Pavlicek, J. | | 登録日 | 2019-03-13 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Discovery of a New Isoxazole-3-hydroxamate-Based Histone Deacetylase 6 Inhibitor SS-208 with Antitumor Activity in Syngeneic Melanoma Mouse Models.
J.Med.Chem., 62, 2019
|
|
6ZW1
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | 分子名称: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | 著者 | Barinka, C, Motlova, L, Ustinova, K. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
