2A6T
 
 | |
2QKM
 
 | |
2QKL
 
 | |
1Q67
 
 | | Crystal structure of Dcp1p | | 分子名称: | Decapping protein involved in mRNA degradation-Dcp1p | | 著者 | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | 登録日 | 2003-08-12 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | 分子名称: | DNA repair protein RAD5, ZINC ION | | 著者 | Shen, M, Xiang, S. | | 登録日 | 2019-11-06 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
3BV4
 
 | | Crystal structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant | | 分子名称: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase A, SULFATE ION | | 著者 | Sherawat, M, Tolan, D.R, Allen, K.N. | | 登録日 | 2008-01-04 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6L8O
 
 | |
5NPQ
 
 | | Human N-myristoyltransferase 1 (MNT1) with Myristoyl-CoA analogue X10 bound | | 分子名称: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, MAGNESIUM ION, ... | | 著者 | Shen, M, Perez-Dorado, I, Fedoryshchak, R, Tate, E.W. | | 登録日 | 2017-04-18 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.372 Å) | | 主引用文献 | Human N-myristoyltransferase 1 (MNT1) with Myristoyl-CoA analogue X10 bound.
To be published
|
|
5CYL
 
 | |
7EIM
 
 | |
7EJT
 
 | |
7EJP
 
 | |
7EKX
 
 | |
7EKW
 
 | |
7EKU
 
 | |
4LMO
 
 | |
4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | 分子名称: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | 著者 | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | 登録日 | 2015-05-01 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
2AYW
 
 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | 著者 | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | 登録日 | 2005-09-09 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4GXZ
 
 | | Crystal structure of a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium | | 分子名称: | Suppression of copper sensitivity protein | | 著者 | Shepherd, M, Heras, B, King, G.J, Argente, M.P, Achard, M.E.S, King, N.P, McEwan, A.G, Schembri, M.A. | | 登録日 | 2012-09-04 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Antioxid Redox Signal, 19, 2013
|
|
3J9G
 
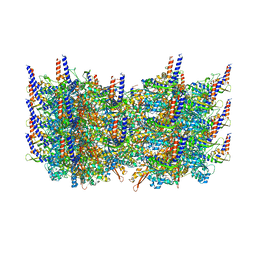 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | 分子名称: | VipA, VipB | | 著者 | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | 登録日 | 2015-01-16 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
1EDO
 
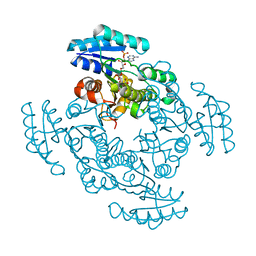 | | THE X-RAY STRUCTURE OF BETA-KETO ACYL CARRIER PROTEIN REDUCTASE FROM BRASSICA NAPUS COMPLEXED WITH NADP+ | | 分子名称: | BETA-KETO ACYL CARRIER PROTEIN REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Fisher, M, Kroon, J.T, Martindale, W, Stuitje, A.R, Slabas, A.R, Rafferty, J.B. | | 登録日 | 2000-01-28 | | 公開日 | 2001-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The X-ray structure of Brassica napus beta-keto acyl carrier protein reductase and its implications for substrate binding and catalysis.
Structure Fold.Des., 8, 2000
|
|
3J70
 
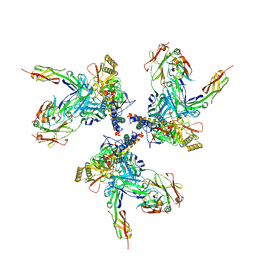 | | Model of gp120, including variable regions, in complex with CD4 and 17b | | 分子名称: | Envelope glycoprotein gp120, T-cell surface glycoprotein CD4, envelope glycoprotein gp41, ... | | 著者 | Rasheed, M, Bettadapura, R, Bajaj, C. | | 登録日 | 2014-04-22 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Computational Refinement and Validation Protocol for Proteins with Large Variable Regions Applied to Model HIV Env Spike in CD4 and 17b Bound State.
Structure, 23, 2015
|
|
6FCO
 
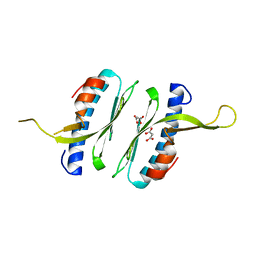 | | Structural and functional characterisation of Frataxin (FXN) like protein from Chaetomium thermophilum | | 分子名称: | MALONIC ACID, Mitochondrial frataxin-like protein | | 著者 | Jamshidiha, M, Rasheed, M, Pastore, A, Cota, E. | | 登録日 | 2017-12-20 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural and functional characterization of a frataxin from a thermophilic organism.
FEBS J., 286, 2019
|
|
7VW7
 
 | | Crystal structure of the 2 ADP-AlF4-bound V1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Shekhar, M, Gupta, C, Singharoy, A, Murata, T. | | 登録日 | 2021-11-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.818 Å) | | 主引用文献 | Revealing a Hidden Intermediate of Rotatory Catalysis with X-ray Crystallography and Molecular Simulations.
Acs Cent.Sci., 8, 2022
|
|
2WY4
 
 | | Structure of bacterial globin from Campylobacter jejuni at 1.35 A resolution | | 分子名称: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SINGLE DOMAIN HAEMOGLOBIN | | 著者 | Barynin, V.V, Sedelnikova, S.E, Shepherd, M, Wu, G, Poole, R.K, Rice, D.W. | | 登録日 | 2009-11-11 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The Single-Domain Globin from the Pathogenic Bacterium Campylobacter Jejuni: Novel D-Helix Conformation, Proximal Hydrogen Bonding that Influences Ligand Binding, and Peroxidase-Like Redox Properties.
J.Biol.Chem., 285, 2010
|
|
