1EHY
 
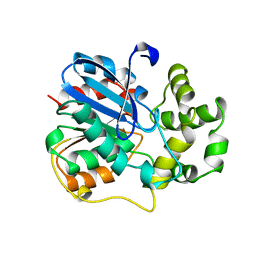 | | X-ray structure of the epoxide hydrolase from agrobacterium radiobacter ad1 | | 分子名称: | POTASSIUM ION, PROTEIN (SOLUBLE EPOXIDE HYDROLASE) | | 著者 | Nardini, M, Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Rink, R, Janssen, D.B, Dijkstra, B.W. | | 登録日 | 1998-10-17 | | 公開日 | 1999-10-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides.
J.Biol.Chem., 274, 1999
|
|
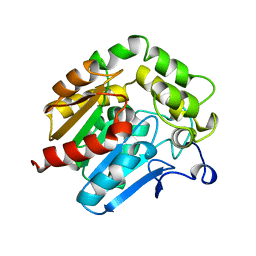
1HDE
 
 | |
2H1Z
 
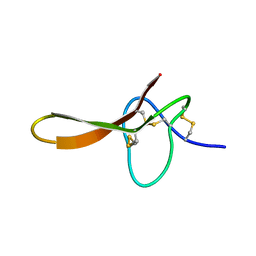 | |
7MSQ
 
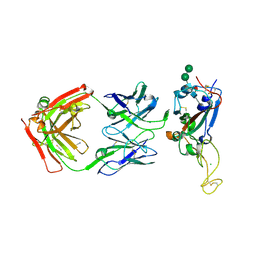 | |
7KZB
 
 | |
7KZC
 
 | |
7KZA
 
 | |
6H9O
 
 | |
6H9N
 
 | |
7PZC
 
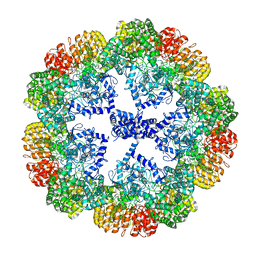 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | 分子名称: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | 著者 | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | 登録日 | 2021-10-12 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
6P4B
 
 | | HyHEL10 fab variant HyHEL10-4x (heavy chain mutations L4F, Y33H, S56N, and Y58F) bound to hen egg lysozyme variant HEL2x-flex (mutations R21Q, R73E, C76S, and C94S) | | 分子名称: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain, ... | | 著者 | Langley, D.B, Christ, D. | | 登録日 | 2019-05-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4C
 
 | | HyHEL10 Fab carrying four heavy chain mutations (HyHEL10-4x): L4F, Y33H, S56N, and Y58F | | 分子名称: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain | | 著者 | Langley, D.B, Christ, D. | | 登録日 | 2019-05-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4D
 
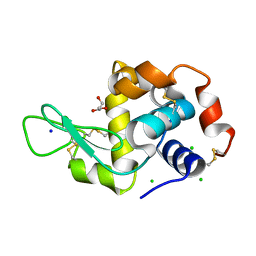 | | Hen egg lysozyme (HEL) containing three point mutations (HEL3x): R21Q, R73E, and D101R | | 分子名称: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | 著者 | Langley, D.B, Christ, D. | | 登録日 | 2019-05-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4A
 
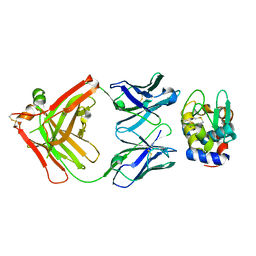 | |
5V8G
 
 | | Pekin duck lysozyme isoform I (DEL-I) | | 分子名称: | CHLORIDE ION, THIOCYANATE ION, lysozyme isoform I | | 著者 | Langley, D.B, Christ, D. | | 登録日 | 2017-03-22 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5V3B
 
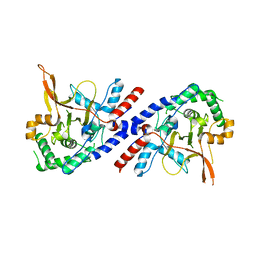 | |
5V92
 
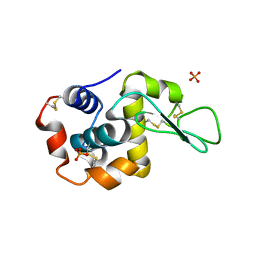 | |
5V94
 
 | | Pekin duck egg lysozyme isoform III (DEL-III), cubic form | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | 著者 | Langley, D.B, Christ, D. | | 登録日 | 2017-03-22 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5V3P
 
 | |
5VAS
 
 | | Pekin duck egg lysozyme isoform III (DEL-III), orthorhombic form | | 分子名称: | GLYCEROL, Lysozyme, PHOSPHATE ION | | 著者 | Christie, M, Christ, D, Langley, D.B. | | 登録日 | 2017-03-27 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5VJO
 
 | |
5VJQ
 
 | | Complex between HyHEL10 Fab fragment heavy chain mutant (I29F, S52T, Y53F) and Pekin duck egg lysozyme isoform I (DEL-I) | | 分子名称: | CHLORIDE ION, GLYCEROL, HyHEL10 heavy chain Fab fragment carrying three mutations; I29F, ... | | 著者 | Langley, D.B, Christ, D. | | 登録日 | 2017-04-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination.
Science, 360, 2018
|
|
