4NPP
 
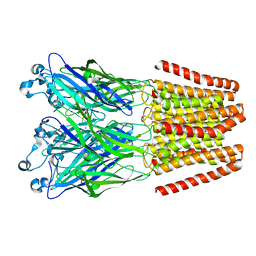 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | 分子名称: | NICKEL (II) ION, Proton-gated ion channel | | 著者 | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | 登録日 | 2013-11-22 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NPQ
 
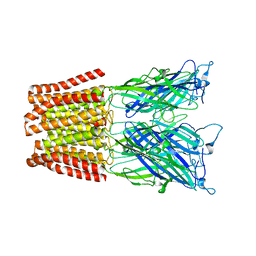 | | The resting-state conformation of the GLIC ligand-gated ion channel | | 分子名称: | Proton-gated ion channel | | 著者 | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | 登録日 | 2013-11-22 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (4.35 Å) | | 主引用文献 | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8SIL
 
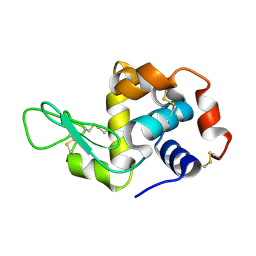 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | 分子名称: | Lysozyme C | | 著者 | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L, Botha, S, Poitevin, F, Sierra, R.G. | | 登録日 | 2023-04-16 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4PIR
 
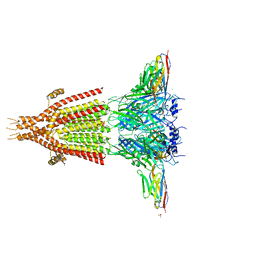 | | X-ray structure of the mouse serotonin 5-HT3 receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | 著者 | Hassaine, G, Deluz, C, Grasso, L, Wyss, R, Tol, M.B, Hovius, R, Graff, A, Stahlberg, H, Tomizaki, T, Desmyter, A, Moreau, C, Li, X.-D, Poitevin, F, Vogel, H, Nury, H. | | 登録日 | 2014-05-09 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | X-ray structure of the mouse serotonin 5-HT3 receptor.
Nature, 512, 2014
|
|
7L9S
 
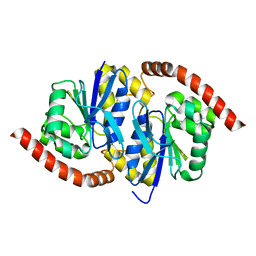 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-04 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9W
 
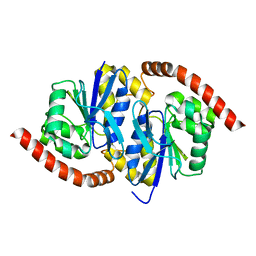 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-3) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-05 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.199 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA0
 
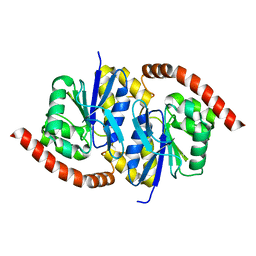 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-2) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-05 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Q
 
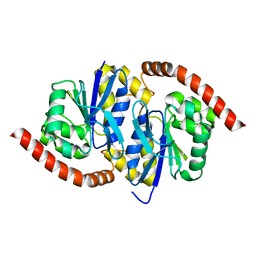 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-04 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.149 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Z
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-1) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-05 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA3
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-3) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-05 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAV
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-1) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-06 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.149 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAX
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-2) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-07 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.198 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LB9
 
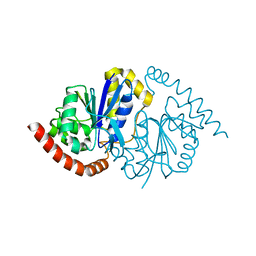 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-3) at 274K, Refmac5-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-07 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LBI
 
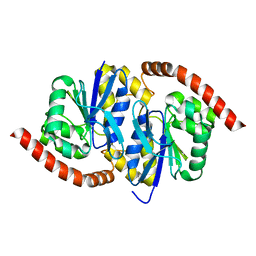 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, PHENIX-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-08 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LBH
 
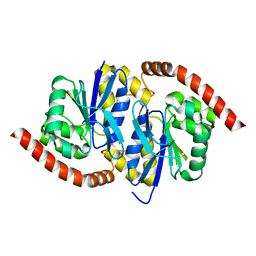 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, PHENIX-refined | | 分子名称: | Isonitrile hydratase InhA | | 著者 | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | 登録日 | 2021-01-08 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
6CAS
 
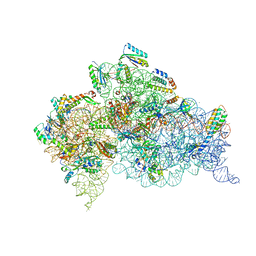 | |
6CAR
 
 | | Serial Femtosecond X-ray Crystal Structure of 30S ribosomal subunit from Thermus thermophilus in complex with Sisomicin | | 分子名称: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | 著者 | DeMirci, H. | | 登録日 | 2018-01-31 | | 公開日 | 2018-07-25 | | 最終更新日 | 2018-10-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Aminoglycoside ribosome interactions reveal novel conformational states at ambient temperature.
Nucleic Acids Res., 46, 2018
|
|
6CAO
 
 | |
3LSV
 
 | |
4HFI
 
 | | The GLIC pentameric Ligand-Gated Ion Channel at 2.4 A resolution | | 分子名称: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | 著者 | Sauguet, L, Corringer, P.J, Delarue, M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4QZA
 
 | | Mouse Tdt in complex with a DSB substrate, C-C base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4QZD
 
 | | Mouse Tdt, F405A mutant, in complex with a DSB substrate, C-C base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4QZH
 
 | | Mouse Tdt, F401A mutant, in complex with a DSB substrate, C-T base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*T)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
7CWB
 
 | |
7CWC
 
 | |
