1KIX
 
 | |
4WFZ
 
 | |
4WFY
 
 | |
4WFX
 
 | |
4ZPB
 
 | |
4ZPD
 
 | |
4ZP8
 
 | |
4ZP6
 
 | |
4ZP9
 
 | |
4ZPA
 
 | |
4ZP7
 
 | |
4ZPC
 
 | |
5VKY
 
 | |
2ILY
 
 | |
2IM2
 
 | |
1TQL
 
 | | POLIOVIRUS POLYMERASE G1A MUTANT | | 分子名称: | ACETIC ACID, RNA-directed RNA polymerase | | 著者 | Thompson, A.A, Peersen, O.B. | | 登録日 | 2004-06-17 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase
Embo J., 23, 2004
|
|
3DDK
 
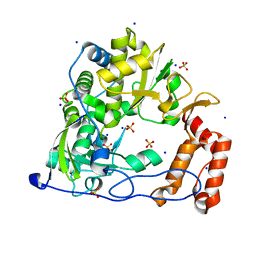 | | Coxsackievirus B3 3Dpol RNA Dependent RNA Polymerase | | 分子名称: | RNA polymerase B3 3Dpol, SODIUM ION, SULFATE ION | | 著者 | Campagnola, G, Weygandt, M.H, Scoggin, K.E, Peersen, O.B. | | 登録日 | 2008-06-05 | | 公開日 | 2008-09-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of Coxsackievirus B3 3Dpol Highlights Functional Importance of Residue 5 in Picornaviral Polymerases
J.Virol., 82, 2008
|
|
1RA7
 
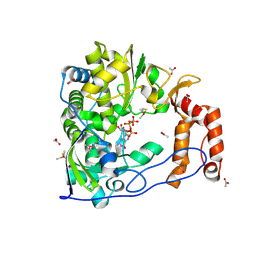 | | Poliovirus Polymerase with GTP | | 分子名称: | ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | 著者 | Thompson, A.A, Peersen, O.B. | | 登録日 | 2003-10-31 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase.
Embo J., 23, 2004
|
|
1RAJ
 
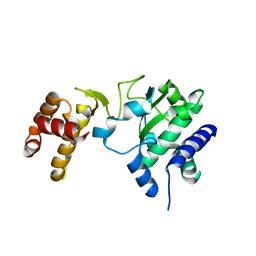 | |
1RA6
 
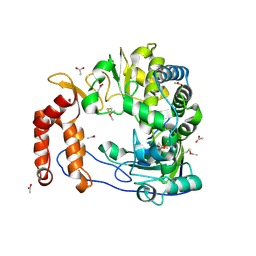 | |
2ILZ
 
 | |
2IM0
 
 | |
2IM1
 
 | |
2IM3
 
 | |
3EVF
 
 | | Crystal structure of Me7-GpppA complex of yellow fever virus methyltransferase and S-adenosyl-L-homocysteine | | 分子名称: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Thompson, A.A, Geiss, B.J, Peersen, O.B. | | 登録日 | 2008-10-13 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Analysis of flavivirus NS5 methyltransferase cap binding.
J.Mol.Biol., 385, 2009
|
|
