7OMT
 
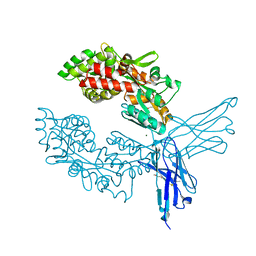 | | Crystal structure of ProMacrobody 21 with bound maltose | | 分子名称: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | 著者 | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | 登録日 | 2021-05-24 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7OMM
 
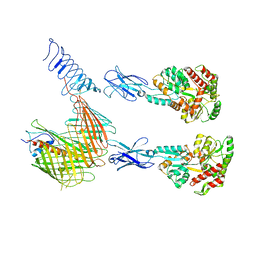 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | 分子名称: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | 著者 | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | 登録日 | 2021-05-24 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | 分子名称: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | 著者 | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | 登録日 | 2021-09-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
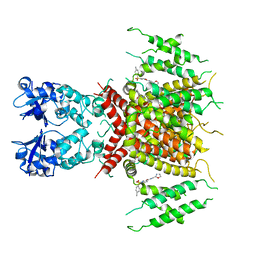 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | 分子名称: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | 著者 | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | 登録日 | 2021-09-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7AA5
 
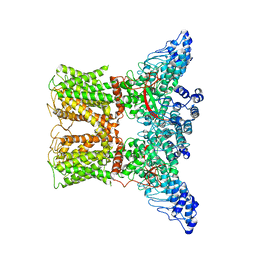 | | Human TRPV4 structure in presence of 4a-PDD | | 分子名称: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | 著者 | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | 登録日 | 2020-09-03 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
1MOU
 
 | | Crystal structure of Coral pigment | | 分子名称: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | 著者 | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | 登録日 | 2002-09-10 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
1MOV
 
 | | Crystal structure of Coral protein mutant | | 分子名称: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | 著者 | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | 登録日 | 2002-09-10 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
3IF9
 
 | | Crystal structure of Glycine Oxidase G51S/A54R/H244A mutant in complex with inhibitor glycolate | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCOLIC ACID, Glycine oxidase | | 著者 | Pedotti, M, Rosini, E, Molla, G, Moschetti, T, Vallone, B, Savino, C, Pollegioni, L. | | 登録日 | 2009-07-24 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Glyphosate resistance by engineering the flavoenzyme glycine oxidase.
J.Biol.Chem., 284, 2009
|
|
2VDC
 
 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | 分子名称: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | 登録日 | 2007-10-04 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
1OCA
 
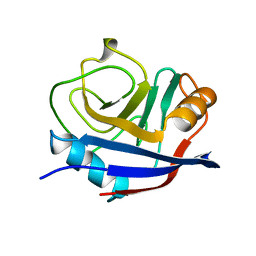 | | HUMAN CYCLOPHILIN A, UNLIGATED, NMR, 20 STRUCTURES | | 分子名称: | CYCLOPHILIN A | | 著者 | Ottiger, M, Zerbe, O, Guntert, P, Wuthrich, K. | | 登録日 | 1997-07-07 | | 公開日 | 1997-11-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution conformation of unligated human cyclophilin A.
J.Mol.Biol., 272, 1997
|
|
1ERD
 
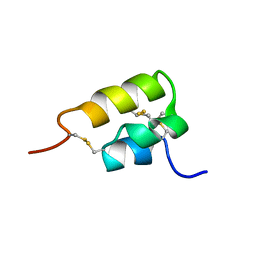 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | 分子名称: | PHEROMONE ER-2 | | 著者 | Ottiger, M, Szyperski, T, Luginbuhl, P, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | 登録日 | 1994-02-14 | | 公開日 | 1994-10-15 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
5MG3
 
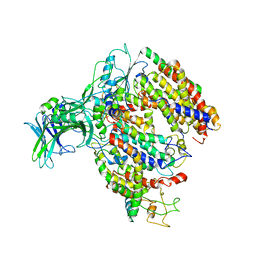 | | EM fitted model of bacterial holo-translocon | | 分子名称: | Membrane protein insertase YidC, Protein translocase subunit SecD, Protein translocase subunit SecE, ... | | 著者 | Schaffitzel, C, Botte, M. | | 登録日 | 2016-11-20 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion.
Sci Rep, 6, 2016
|
|
6QGO
 
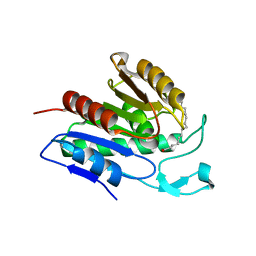 | | Crystal structure of APT1 S119A mutant bound to palmitic acid. | | 分子名称: | Acyl-protein thioesterase 1, PALMITIC ACID | | 著者 | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | 登録日 | 2019-01-12 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.599 Å) | | 主引用文献 | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6QGQ
 
 | | Crystal structure of APT1 C2S mutant bound to palmitic acid. | | 分子名称: | Acyl-protein thioesterase 1, GLYCEROL, PALMITIC ACID | | 著者 | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | 登録日 | 2019-01-12 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6QGS
 
 | | Crystal structure of APT1 bound to palmitic acid. | | 分子名称: | Acyl-protein thioesterase 1, CHLORIDE ION, PALMITIC ACID | | 著者 | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | 登録日 | 2019-01-12 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.755 Å) | | 主引用文献 | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6AXD
 
 | |
5OJA
 
 | | Structure of MbQ | | 分子名称: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | 登録日 | 2017-07-21 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.347 Å) | | 主引用文献 | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJB
 
 | | Structure of MbQ NMH | | 分子名称: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | 登録日 | 2017-07-21 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.543 Å) | | 主引用文献 | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJC
 
 | | Structure of MbQ2.1 NMH | | 分子名称: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | 登録日 | 2017-07-21 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJ9
 
 | | Structure of Mb NMH | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | 登録日 | 2017-07-21 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.483 Å) | | 主引用文献 | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
6RWT
 
 | | Crystal structure of the Cbp3 homolog from Brucella abortus | | 分子名称: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Masuyer, G, Ndi, M, Ott, M, Stenmark, P. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structural basis for the interaction of the chaperone Cbp3 with newly synthesized cytochromebduring mitochondrial respiratory chain assembly.
J.Biol.Chem., 294, 2019
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | 分子名称: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
6GIQ
 
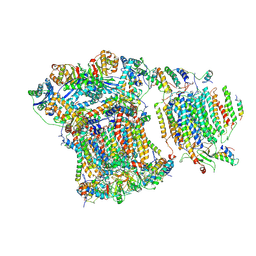 | | Saccharomyces cerevisiae respiratory supercomplex III2IV | | 分子名称: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | 著者 | Rathore, S, Berndtsson, J, Conrad, J, Ott, M. | | 登録日 | 2018-05-15 | | 公開日 | 2019-01-02 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Cryo-EM structure of the yeast respiratory supercomplex.
Nat. Struct. Mol. Biol., 26, 2019
|
|
