2QA1
 
 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | 登録日 | 2007-06-14 | | 公開日 | 2007-08-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QA2
 
 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | 著者 | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | 登録日 | 2007-06-14 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
3KG1
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | 分子名称: | CHLORIDE ION, SnoaB | | 著者 | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | 登録日 | 2009-10-28 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG0
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | 著者 | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | 登録日 | 2009-10-28 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KNG
 
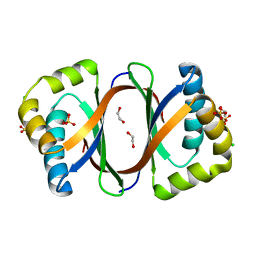 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | 著者 | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | 登録日 | 2009-11-12 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
5ZUU
 
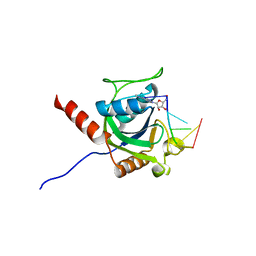 | | Crystal structure of AtCPSF30 YTH domain in complex with 10mer m6A-modified RNA | | 分子名称: | 30-kDa cleavage and polyadenylation specificity factor 30, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Wu, B, Nie, H, Li, S, Patel, D.J. | | 登録日 | 2018-05-08 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis.
Mol Plant, 2021
|
|
4V4D
 
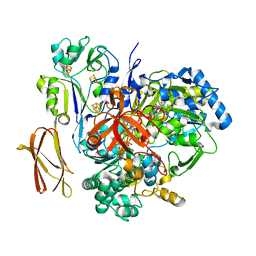 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with pyrogallol | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,3-TRIOL, CALCIUM ION, ... | | 著者 | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | 登録日 | 2004-06-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
3UVC
 
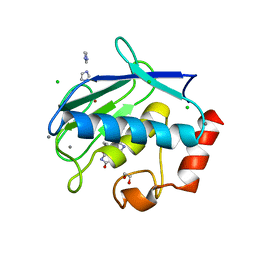 | | MMP12 in a complex with the dimeric adduct: 5-(5-phenylhydantoin)-5-phenylhydantoin | | 分子名称: | (4R,4'S)-4,4'-diphenyl-4,4'-biimidazolidine-2,2',5,5'-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | 著者 | Derbyshire, D.J, Danielson, H, Nystrum, S. | | 登録日 | 2011-11-29 | | 公開日 | 2013-01-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Characterization of fragments interacting with MMP-12
To be Published
|
|
4P9R
 
 | | Speciation of a group I intron into a lariat capping ribozyme (Heavy atom derivative) | | 分子名称: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (189-MER) | | 著者 | Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E, Masquida, B. | | 登録日 | 2014-04-04 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Speciation of a group I intron into a lariat capping ribozyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4V4E
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | 著者 | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | 登録日 | 2004-06-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4V4C
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, CALCIUM ION, ... | | 著者 | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | 登録日 | 2004-06-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
3TGX
 
 | | IL-21:IL21R complex | | 分子名称: | Interleukin-21, Interleukin-21 receptor, NICKEL (II) ION, ... | | 著者 | Hamming, O.J, Kang, L, Svenson, A, Karlsen, J.L, Rahbek-Nielsen, H, Paludan, S.R, Hjort, S.A, Bondensgaard, K, Hartmann, R. | | 登録日 | 2011-08-18 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of the interleukin 21 receptor bound to interleukin 21 reveals that a sugar chain interacting with the WSXWS motif is an integral part of the interleukin 21 receptor.
J.Biol.Chem., 2012
|
|
1XDS
 
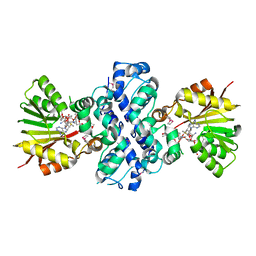 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) and 11-deoxy-beta-rhodomycin (DbrA) | | 分子名称: | 11-DEOXY-BETA-RHODOMYCIN, Protein RdmB, S-ADENOSYLMETHIONINE | | 著者 | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-09-08 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
1XDU
 
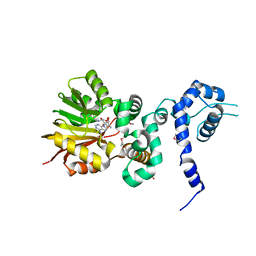 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with Sinefungin (SFG) | | 分子名称: | ACETATE ION, Protein RdmB, SINEFUNGIN | | 著者 | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J. | | 登録日 | 2004-09-08 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
1TW3
 
 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | 分子名称: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G. | | 登録日 | 2004-06-30 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
1TW2
 
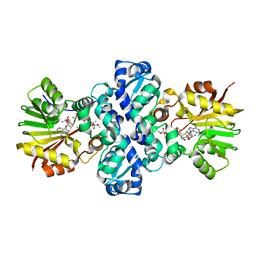 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | 分子名称: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-06-30 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
3IHG
 
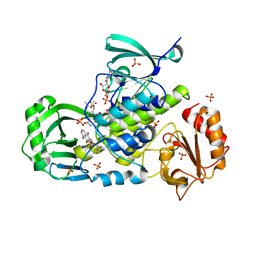 | | Crystal structure of a ternary complex of aklavinone-11 hydroxylase with FAD and aklavinone | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, RdmE, SULFATE ION, ... | | 著者 | Lindqvist, Y, Koskiniemi, H, Jansson, A, Sandalova, T, Schneider, G. | | 登録日 | 2009-07-30 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
J.Mol.Biol., 393, 2009
|
|
4P8Z
 
 | | Speciation of a group I intron into a lariat capping ribozyme | | 分子名称: | Didymium iridis partial IGS, 18S rRNA gene, I-DirI gene and partial ITS1 | | 著者 | Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E, Masquida, B. | | 登録日 | 2014-04-01 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Speciation of a group I intron into a lariat capping ribozyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1XV8
 
 | | Crystal Structure of Human Salivary Alpha-Amylase Dimer | | 分子名称: | Alpha-amylase, CALCIUM ION, CHLORIDE ION | | 著者 | Fisher, S.Z, Govindasamy, L, Tu, C.K, Silverman, D.N, Rajaniemi, H, McKenna, R. | | 登録日 | 2004-10-27 | | 公開日 | 2005-10-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Human Salivary Alpha-Amylase Dimer
To be Published
|
|
6GYV
 
 | | Lariat-capping ribozyme (circular permutation form) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lariat-capping ribozyme, MAGNESIUM ION, ... | | 著者 | Masquida, B, Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E. | | 登録日 | 2018-07-02 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.50003624 Å) | | 主引用文献 | Speciation of a group I intron into a lariat capping ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
2HYB
 
 | | Crystal Structure of Hexameric DsrEFH | | 分子名称: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | 著者 | Shin, D.H, Connie, H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2006-08-04 | | 公開日 | 2007-07-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of hexameric DsrEFH
To be Published
|
|
6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | 分子名称: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | 著者 | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | 登録日 | 2018-12-02 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.749 Å) | | 主引用文献 | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J23
 
 | | Crystal structure of arabidopsis ADAL complexed with GMP | | 分子名称: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | 著者 | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | 登録日 | 2018-12-30 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | 分子名称: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | 著者 | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | 登録日 | 2019-01-10 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
7P58
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 1-[6-(3-propan-2-yloxyphenyl)pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Davies, T.G. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.886 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
