7RNO
 
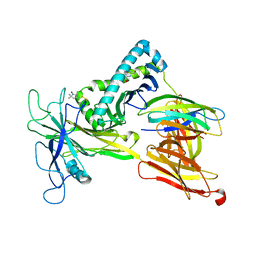 | | Model of the Ac-6-FP/hpMR1/bB2m/TAPBPR complex from integrated docking, NMR and restrained MD | | 分子名称: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | 著者 | McShan, A.C, Sgourakis, N.G. | | 登録日 | 2021-07-29 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-08-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules.
Nat.Chem.Biol., 18, 2022
|
|
8T62
 
 | |
8T61
 
 | |
8T63
 
 | |
8TB1
 
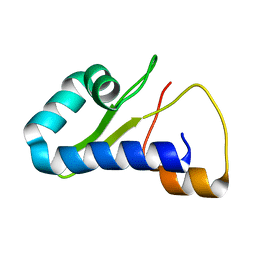 | |
8TV4
 
 | |
8TXS
 
 | |
6NPR
 
 | | Crystal structure of H-2Dd with C84-C139 disulfide in complex with gp120 derived peptide P18-I10 | | 分子名称: | ARG-GLY-PRO-GLY-ARG-ALA-PHE-VAL-THR-ILE, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Toor, J, McShan, A.C, Tripathi, S.M, Sgourakis, N.G. | | 登録日 | 2019-01-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OSW
 
 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | 分子名称: | Forkhead box M1 | | 著者 | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | 登録日 | 2019-05-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
9BVS
 
 | |
9BVU
 
 | |
9BVV
 
 | |
6MPP
 
 | |
6B9K
 
 | |
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | 分子名称: | De novo beta protein | | 著者 | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | 登録日 | 2018-07-19 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5VZ5
 
 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | 著者 | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | 登録日 | 2017-05-26 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5901 Å) | | 主引用文献 | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
5TXS
 
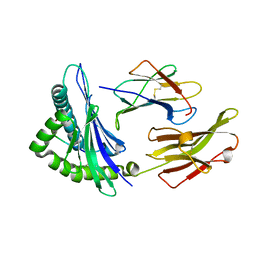 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-15 alpha chain, ... | | 著者 | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | 登録日 | 2016-11-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
7SKN
 
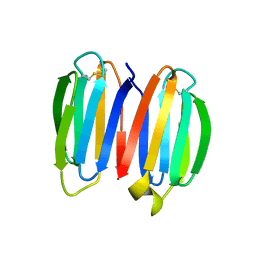 | |
6AT9
 
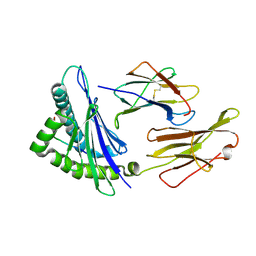 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-A*0101 | | 分子名称: | ALK, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | 登録日 | 2017-08-28 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9503 Å) | | 主引用文献 | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
5IVX
 
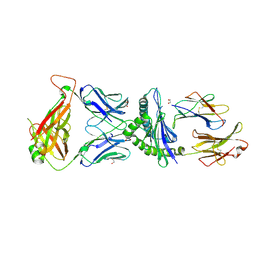 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Natarajan, K, Jiang, J, Margulies, D. | | 登録日 | 2016-03-21 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | 分子名称: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | 著者 | Park, J, Baker, D. | | 登録日 | 2018-12-03 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.039 Å) | | 主引用文献 | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | 分子名称: | Design construct XAA | | 著者 | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | 登録日 | 2019-02-16 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | 分子名称: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | 著者 | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | 主引用文献 | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6NY8
 
 | |
