8WKY
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | 分子名称: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | 著者 | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8WKZ
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | 分子名称: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | 著者 | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
5OCQ
 
 | | Crystal structure of the complex of the kappa-carrageenase from Pseudoalteromonas carrageenovora with an oligotetrasaccharide of kappa-carrageenan | | 分子名称: | 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, CITRIC ACID, ... | | 著者 | Czjzek, M, Leroux, C, Bernard, T, Matard-Mann, M, Jeudy, A, Michel, G. | | 登録日 | 2017-07-03 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
4J19
 
 | | Structure of a novel telomere repeat binding protein bound to DNA | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | 著者 | Kappei, D, Butter, F, Benda, C, Scheibe, M, Draskovic, I, Stevense, M, Lopes Novo, C, Basquin, C, Krastev, D.B, Kittler, R, Jessberger, R, Londono-Vallejo, A.J, Mann, M, Buchholz, F. | | 登録日 | 2013-02-01 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment.
Embo J., 32, 2013
|
|
5OCR
 
 | | Crystal structure of the kappa-carrageenase zobellia_236 from Zobellia galactanivorans | | 分子名称: | GLYCEROL, Kappa-carrageenase, MAGNESIUM ION | | 著者 | Czjzek, M, Matard-Mann, M, Michel, G, Jeudy, A, Larocque, R. | | 登録日 | 2017-07-03 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
2UYZ
 
 | | Non-covalent complex between Ubc9 and SUMO1 | | 分子名称: | SMALL UBIQUITIN-RELATED MODIFIER 1, SODIUM ION, SUMO-CONJUGATING ENZYME UBC9 | | 著者 | Knipscheer, P, van Dijk, W.J, Olsen, J.V, Mann, M, Sixma, T.K. | | 登録日 | 2007-04-21 | | 公開日 | 2007-06-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation.
EMBO J., 26, 2007
|
|
2VRR
 
 | | Structure of SUMO modified Ubc9 | | 分子名称: | FORMIC ACID, SMALL UBIQUITIN-RELATED MODIFIER 1, SODIUM ION, ... | | 著者 | Knipscheer, P, Flotho, A, Klug, H, Olsen, J.V, van Dijk, W.J, Fish, A, Johnson, E.S, Mann, M, Sixma, T.K, Pichler, A. | | 登録日 | 2008-04-13 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Ubc9 sumoylation regulates SUMO target discrimination.
Mol. Cell, 31, 2008
|
|
4W8Z
 
 | |
4W8W
 
 | |
4W8V
 
 | |
4W8X
 
 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | 分子名称: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | 著者 | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | 登録日 | 2014-08-26 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4W8Y
 
 | | Structure of full length Cmr2 from Pyrococcus furiosus (Manganese bound form) | | 分子名称: | CRISPR system Cmr subunit Cmr2, MANGANESE (II) ION, ZINC ION | | 著者 | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | 登録日 | 2014-08-26 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
8CDK
 
 | |
8CDJ
 
 | |
8CAF
 
 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | 分子名称: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | 登録日 | 2023-01-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
4ZTD
 
 | | Crystal Structure of Human PCNA in complex with a TRAIP peptide | | 分子名称: | ALA-GLY-ALA-GLY-ALA, ALA-PHE-GLN-ALA-LYS-LEU-ASP-THR-PHE-LEU-TRP-SER, Proliferating cell nuclear antigen | | 著者 | Montoya, G, Mortuza, G.B, Blanco, F.J, Ibanez de Opakua, A. | | 登録日 | 2015-05-14 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
J.Cell Biol., 212, 2016
|
|
8Q8J
 
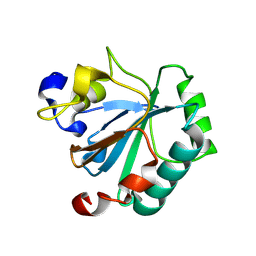 | | Crystal structure of human GPX4-R152H | | 分子名称: | Glutathione peroxidase | | 著者 | Napolitano, V, Mourao, A, Kolonko, M, Bostock, M.J, Sattler, M, Conrad, M, Popowicz, G. | | 登録日 | 2023-08-18 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of human GPX4-R152H
To Be Published
|
|
8Q8N
 
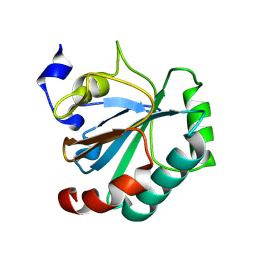 | | Crystal structure of human GPX4-U46C-I129S-L130S | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | 登録日 | 2023-08-18 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
7WUG
 
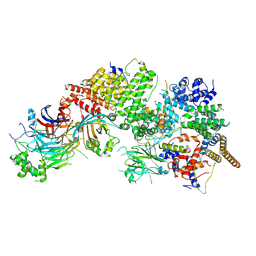 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | 分子名称: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Cheng, J.D, Schulman, B.A. | | 登録日 | 2022-02-08 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
5OPQ
 
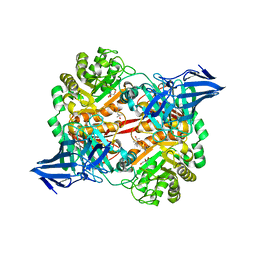 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | 著者 | Ficko-Blean, E, Michel, G, Czjzek, M. | | 登録日 | 2017-08-10 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
5OLC
 
 | |
7NS5
 
 | | Structure of yeast Fbp1 (Fructose-1,6-bisphosphatase 1) | | 分子名称: | Fructose-1,6-bisphosphatase, MAGNESIUM ION, PHOSPHATE ION | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NSC
 
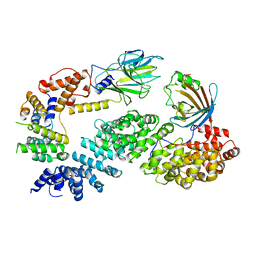 | | Substrate receptor scaffolding module of human CTLH E3 ubiquitin ligase | | 分子名称: | Glucose-induced degradation protein 4 homolog, Glucose-induced degradation protein 8 homolog, Isoform 2 of Armadillo repeat-containing protein 8, ... | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NS4
 
 | | Catalytic module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | 分子名称: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, ZINC ION | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NSB
 
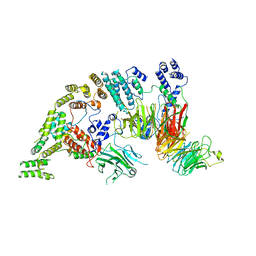 | | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | 分子名称: | Glucose-induced degradation protein 7, Glucose-induced degradation protein 8, Vacuolar import and degradation protein 30 | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
