3ZLW
 
 | | Crystal structure of MEK1 in complex with fragment 3 | | 分子名称: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
 | | Crystal structure of MEK1 in complex with fragment 18 | | 分子名称: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | 分子名称: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZM4
 
 | | Crystal structure of MEK1 in complex with fragment 1 | | 分子名称: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-05 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLY
 
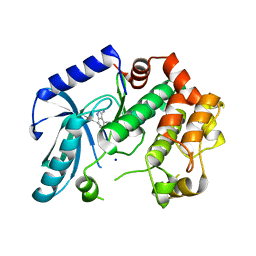 | | Crystal structure of MEK1 in complex with fragment 8 | | 分子名称: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | 著者 | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5LO5
 
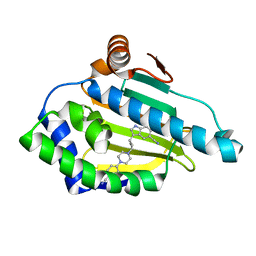 | | HSP90 WITH indole derivative | | 分子名称: | 3-[4-[4-(4-cyanophenyl)piperazin-1-yl]butyl]-6-oxidanyl-1~{H}-indole-5-carbonitrile, Heat shock protein HSP 90-alpha | | 著者 | Graedler, U, Amaral, M. | | 登録日 | 2016-08-08 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
5LO6
 
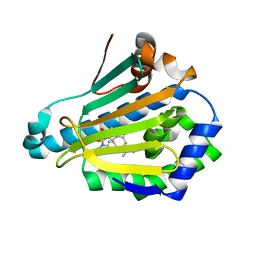 | | HSP90 WITH indazole derivative | | 分子名称: | 3-(3,3-dimethylbutyl)-~{N}-methyl-~{N}-[4-(1-methylpiperidin-4-yl)phenyl]-6-oxidanyl-2~{H}-indazole-5-carboxamide, Heat shock protein HSP 90-alpha | | 著者 | Graedler, U, Amaral, M. | | 登録日 | 2016-08-08 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
5LRL
 
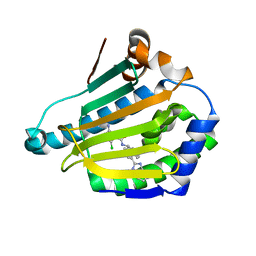 | | CRYSTAL STRUCTURE OF HSP90 IN COMPLEX WITH A003492875 | | 分子名称: | 2-azanyl-5-chloranyl-~{N}-[(9~{R})-4-(1~{H}-imidazo[4,5-c]pyridin-2-yl)-9~{H}-fluoren-9-yl]pyrimidine-4-carboxamide, Heat shock protein HSP 90-alpha | | 著者 | Vallee, F, Dupuy, A. | | 登録日 | 2016-08-19 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations.
J.Chem.Inf.Model., 59, 2019
|
|
5HNI
 
 | | CRYSTAL STRUCTURE OF CMET WT with compound 3 | | 分子名称: | Hepatocyte growth factor receptor, methyl (6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1H-benzimidazol-2-yl)carbamate | | 著者 | Vallee, F, Houtmann, J. | | 登録日 | 2016-01-18 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HO6
 
 | | CRYSTAL STRUCTURE OF CMET IN COMPLEX WITH CMPD. | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Houtmann, J. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOR
 
 | | Crystal structure of c-Met-M1250T in complex with SAR125844. | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Houtmann, J, Marquette, J.-P. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HLW
 
 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | 分子名称: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | 登録日 | 2016-01-15 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOA
 
 | | Crystal structure of c-Met L1195V in complex with SAR125844 | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Marquette, J.-P. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
6ELP
 
 | |
6EI5
 
 | |
7A1X
 
 | | KRASG12C GDP form in complex with Cpd1 | | 分子名称: | 3-(imidazol-1-ylmethyl)-1~{H}-indole, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Mathieu, M, Steier, V. | | 登録日 | 2020-08-14 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | KRAS G12C fragment screening renders new binding pockets.
Small Gtpases, 13, 2022
|
|
7A1W
 
 | | KRASG12C GDP form in complex with Cpd3 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | 著者 | Mathieu, M, Steier, V. | | 登録日 | 2020-08-14 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | KRAS G12C fragment screening renders new binding pockets.
Small Gtpases, 13, 2022
|
|
7A1Y
 
 | |
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-09-29 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6ELO
 
 | |
6EL5
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EY9
 
 | |
6EY8
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha, SULFATE ION, ... | | 著者 | Musil, D, Lehmann, M, Buchstaller, H.-P. | | 登録日 | 2017-11-11 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EYA
 
 | |
6EYB
 
 | |
