2X1J
 
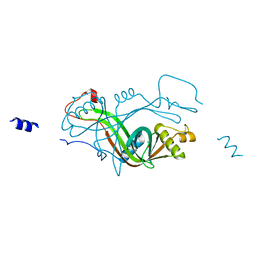 | |
2X1K
 
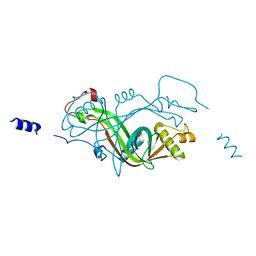 | |
1O7D
 
 | | The structure of the bovine lysosomal a-mannosidase suggests a novel mechanism for low pH activation | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Heikinheimo, P, Helland, R, Leiros, H.S, Leiros, I, Karlsen, S, Evjen, G, Ravelli, R, Schoehn, G, Ruigrok, R, Tollersrud, O.-K, Mcsweeney, S, Hough, E. | | 登録日 | 2002-10-30 | | 公開日 | 2003-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The Structure of Bovine Lysosomal Alpha-Mannosidase Suggests a Novel Mechanism for Low-Ph Activation
J.Mol.Biol., 327, 2003
|
|
4UWR
 
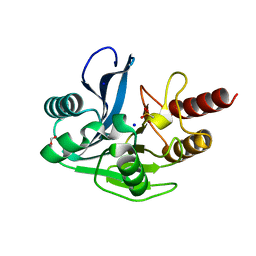 | | Mono-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | 分子名称: | FORMIC ACID, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | 著者 | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | 登録日 | 2014-08-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4UWS
 
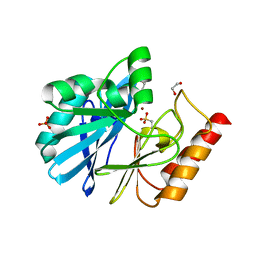 | | VIM-26-PEG. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, METALLO-BETA-LACTAMASE VIM-26, ... | | 著者 | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | 登録日 | 2014-08-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4UWP
 
 | | Penta Zn1 coordination. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | 分子名称: | METALLO-BETA-LACTAMASE VIM-26, ZINC ION | | 著者 | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | 登録日 | 2014-08-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4UWO
 
 | | Native di-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | 分子名称: | GLYCEROL, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | 著者 | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | 登録日 | 2014-08-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.555 Å) | | 主引用文献 | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4BPT
 
 | |
6SKP
 
 | |
6SKR
 
 | | OXA-10_ETP. Structural insight to the enhanced carbapenem efficiency of OXA-655 compared to OXA-10. | | 分子名称: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, FORMIC ACID, ... | | 著者 | Leiros, H.-K.S. | | 登録日 | 2019-08-16 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10.
Febs Open Bio, 10, 2020
|
|
6SKQ
 
 | |
7OBN
 
 | |
8BHX
 
 | |
6RAU
 
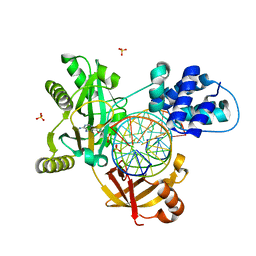 | | PostS3_Pmar_lig4_WT | | 分子名称: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA (5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*CP*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3'), ... | | 著者 | Leiros, H.K.S, Williamson, A. | | 登録日 | 2019-04-08 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
6RAS
 
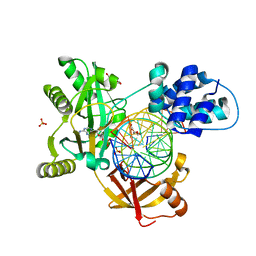 | | Pmar-Lig_Pre. | | 分子名称: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA, ... | | 著者 | Leiros, H.K.S, Williamson, A. | | 登録日 | 2019-04-07 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
6RAR
 
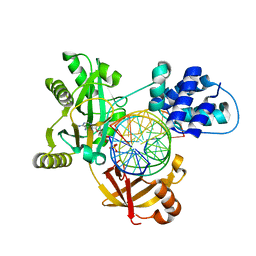 | | Pmar-Lig_PreS3-Mn | | 分子名称: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA (5'-D(*TP*TP*CP*CP*GP*AP*CP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | 著者 | Leiros, H.K.S, Williamson, A. | | 登録日 | 2019-04-07 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.785 Å) | | 主引用文献 | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
6RCE
 
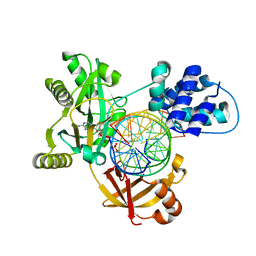 | | Pmar-Lig_PreS3 | | 分子名称: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA (5'-D(*TP*TP*CP*CP*GP*AP*CP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | 著者 | Leiros, H.K.S, Williamson, A. | | 登録日 | 2019-04-11 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.946 Å) | | 主引用文献 | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
6GDR
 
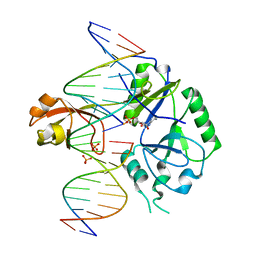 | | DNA binding with a minimal scaffold: Structure-function analysis of Lig E DNA ligases | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | 著者 | Williamson, A, Grigic, M, Leiros, H.K.S. | | 登録日 | 2018-04-24 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases.
Nucleic Acids Res., 46, 2018
|
|
6GOA
 
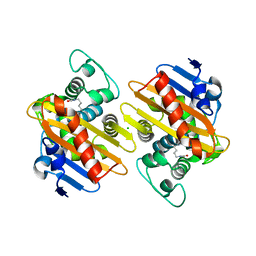 | |
4V2I
 
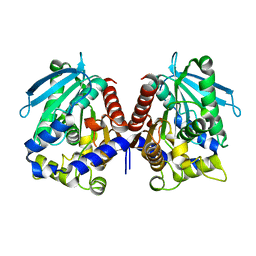 | | Biochemical characterization and structural analysis of a new cold- active and salt tolerant esterase from the marine bacterium Thalassospira sp | | 分子名称: | ESTERASE/LIPASE, MAGNESIUM ION | | 著者 | Santi, C.D, Leiros, H.-K.S, Scala, A.D, Pascale, D.D, Altermark, B, Willassen, N.-P. | | 登録日 | 2014-10-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.686 Å) | | 主引用文献 | Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
Extremophiles, 20, 2016
|
|
6Q5F
 
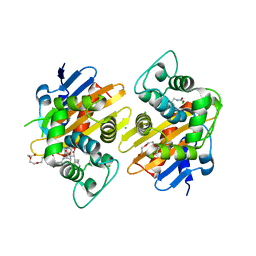 | | OXA-48_P68A-CAZ. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | 分子名称: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.-K.S, Samuelsen, O. | | 登録日 | 2018-12-08 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
6Q5B
 
 | | OXA-48_P68A-AVI. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CARBON DIOXIDE, ... | | 著者 | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.K.S, Samuelsen, O. | | 登録日 | 2018-12-07 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
8PEB
 
 | |
8PEC
 
 | | OXA-48_Q5-CAZ. Epistasis Arises from Shifting the Rate-Limiting Step during Enzyme Evolution | | 分子名称: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION | | 著者 | Leiros, H.-K.S, Frohlich, C. | | 登録日 | 2023-06-13 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Epistasis arises from shifting the rate-limiting step during enzyme evolution of a beta-lactamase.
Nat Catal, 7, 2024
|
|
8PEA
 
 | |
