6OIS
 
 | | CryoEM structure of Arabidopsis DR complex (DMS3-RDM1) | | 分子名称: | Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | 著者 | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
6OIT
 
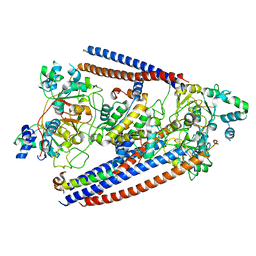 | | CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1) | | 分子名称: | Protein CHROMATIN REMODELING 35, Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | 著者 | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
8HIL
 
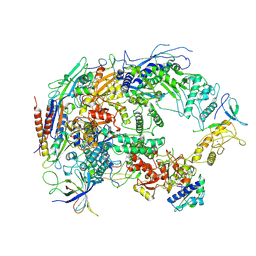 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | 分子名称: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | 著者 | Du, X, Xie, G, Hu, H, Du, J. | | 登録日 | 2022-11-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8HIM
 
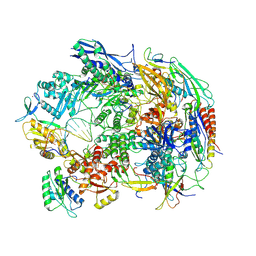 | | A cryo-EM structure of B. oleracea RNA polymerase V elongation complex at 2.73 Angstrom | | 分子名称: | DNA (34-MER), DNA-directed RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, ... | | 著者 | Hu, H, Xie, G, Du, X, Du, J. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8DC2
 
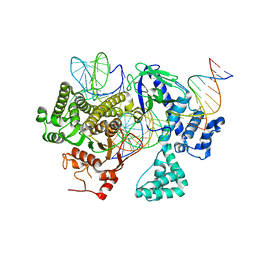 | | Cryo-EM structure of CasLambda (Cas12l) bound to crRNA and DNA | | 分子名称: | CasLambda, DNA NTS, DNA TS, ... | | 著者 | Al-Shayeb, B, Skopintsev, P, Soczek, K, Doudna, J. | | 登録日 | 2022-06-15 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors.
Cell, 185, 2022
|
|
7VG2
 
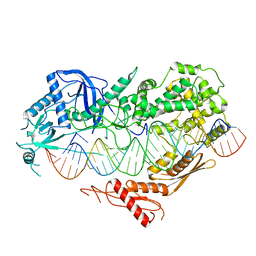 | |
7VG3
 
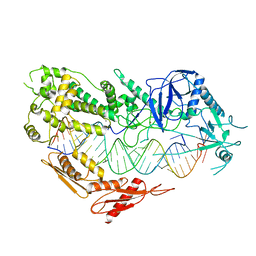 | |
5HH7
 
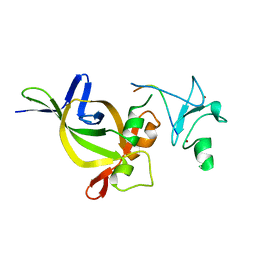 | |
5IX2
 
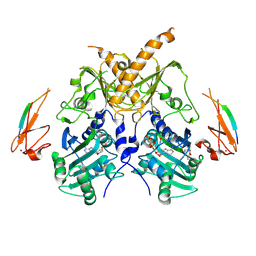 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and unmodified H3 peptide | | 分子名称: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Du, J, Patel, D.J. | | 登録日 | 2016-03-23 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QEP
 
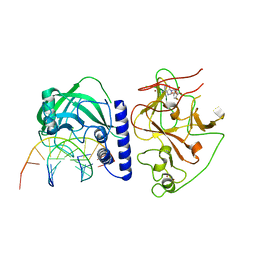 | | crystal structure of KRYPTONITE in complex with mCHG DNA and SAH | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
4QEO
 
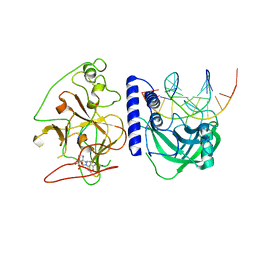 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | 分子名称: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
3Q0B
 
 | |
3Q0F
 
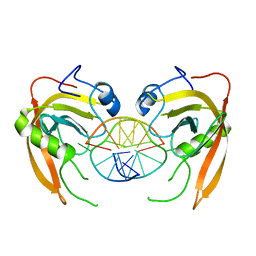 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | 分子名称: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3Q0C
 
 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | 分子名称: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6567 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3Q0D
 
 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3704 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
2ZO0
 
 | | mouse NP95 SRA domain DNA specific complex 1 | | 分子名称: | DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), E3 ubiquitin-protein ligase UHRF1 | | 著者 | Hashimoto, H, Horton, J.R, Cheng, X. | | 登録日 | 2008-05-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
4ONJ
 
 | |
2ZO1
 
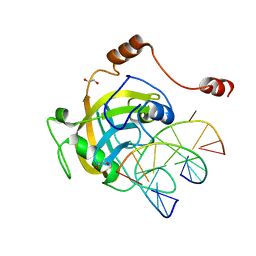 | | Mouse NP95 SRA domain DNA specific complex 2 | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), ... | | 著者 | Hashimoto, H, Horton, J.R, Cheng, X. | | 登録日 | 2008-05-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
4NJ5
 
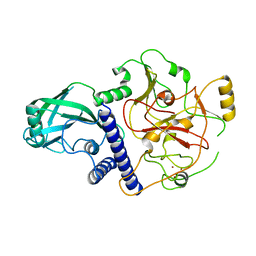 | | Crystal structure of SUVH9 | | 分子名称: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | 著者 | Du, J, Patel, D.J. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
2ZO2
 
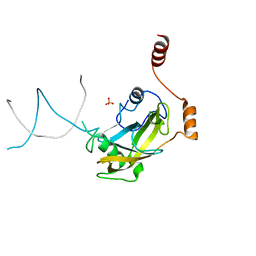 | |
4ONQ
 
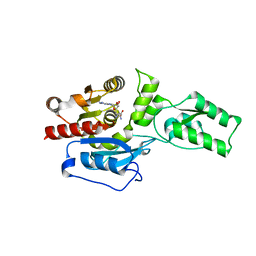 | |
4QEN
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA and SAH | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*TP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
5ZE4
 
 | | The structure of holo- structure of DHAD complex with [2Fe-2S] cluster | | 分子名称: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | 著者 | Zhou, J, Zang, X, Tang, Y, Yan, Y, Gan, J, Wu, L. | | 登録日 | 2018-02-26 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action.
Nature, 559, 2018
|
|
6A5N
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | 分子名称: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Li, X, Du, J. | | 登録日 | 2018-06-24 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
 | |
