8JLW
 
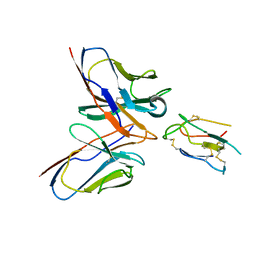 | | CCHFV envelope protein Gc in complex with Gc8 | | 分子名称: | Glycoprotein C,CCHFV envelope protein Gc fusion loops, Mouse antibody Gc8 heavy chain, Mouse antibody Gc8 light chain | | 著者 | Chong, T, Cao, S. | | 登録日 | 2023-06-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
8JKD
 
 | |
8JLX
 
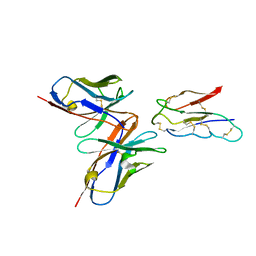 | | CCHFV envelope protein Gc in complex with Gc13 | | 分子名称: | Glycoprotein C,CCHFV Gc fusion loops, Mouse antibody Gc13 heavy chain, Mouse antibody Gc13 light chain | | 著者 | Chong, T, Cao, S. | | 登録日 | 2023-06-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
4ZS3
 
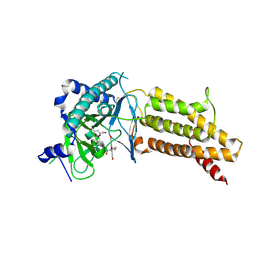 | | Structural complex of 5-aminofluorescein bound to the FTO protein | | 分子名称: | 2-OXOGLUTARIC ACID, 5-amino-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | 著者 | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | 登録日 | 2015-05-13 | | 公開日 | 2015-11-04 | | 最終更新日 | 2015-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
4ZS2
 
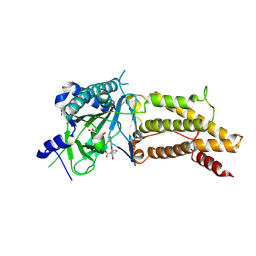 | | Structural complex of FTO/fluorescein | | 分子名称: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | 著者 | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | 登録日 | 2015-05-13 | | 公開日 | 2015-11-04 | | 最終更新日 | 2015-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
1SSZ
 
 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | 分子名称: | Pulmonary surfactant-associated protein B | | 著者 | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | 登録日 | 2004-03-24 | | 公開日 | 2004-06-15 | | 最終更新日 | 2019-04-24 | | 実験手法 | INFRARED SPECTROSCOPY | | 主引用文献 | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
1FRY
 
 | | THE SOLUTION STRUCTURE OF SHEEP MYELOID ANTIMICROBIAL PEPTIDE, RESIDUES 1-29 (SMAP29) | | 分子名称: | MYELOID ANTIMICROBIAL PEPTIDE | | 著者 | Tack, B.F, Sawai, M.V, Kearney, W.R, Robertson, A.D, Sherman, M.A, Wang, W, Hong, T, Boo, L.M, Wu, H, Waring, A.J, Lehrer, R.I. | | 登録日 | 2000-09-07 | | 公開日 | 2002-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SMAP-29 has two LPS-binding sites and a central hinge.
Eur.J.Biochem., 269, 2002
|
|
5ZYR
 
 | | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii | | 分子名称: | ACETATE ION, FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, ... | | 著者 | Oonanant, W, Phongsak, T, Sucharitakul, J, Chaiyen, P, Yuvaniyama, J. | | 登録日 | 2018-05-28 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.20001316 Å) | | 主引用文献 | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii
To Be Published
|
|
8WMC
 
 | |
8WMI
 
 | |
8WM4
 
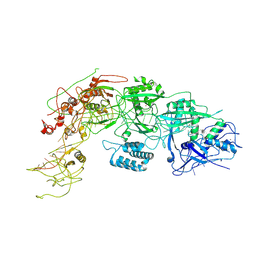 | |
8WML
 
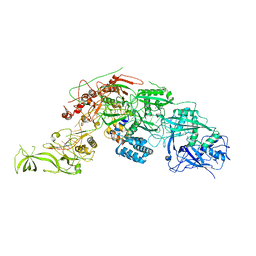 | |
5Y08
 
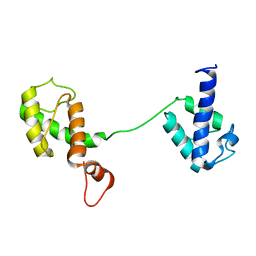 | |
8B30
 
 | |
6UEW
 
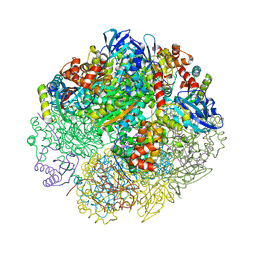 | |
4C13
 
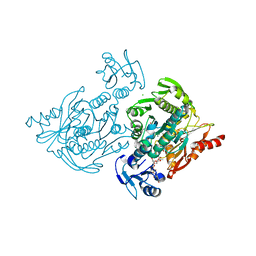 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | 登録日 | 2013-08-09 | | 公開日 | 2013-10-02 | | 最終更新日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
8W9Q
 
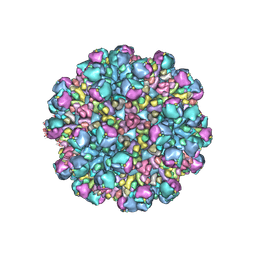 | | Structure of partial Banna virus | | 分子名称: | VP10, VP2, VP4, ... | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-09-05 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8W9R
 
 | | Structure of Banna virus core | | 分子名称: | VP10, VP8, Vp2 | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-09-05 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8W9P
 
 | | Structure of full Banna virus | | 分子名称: | VP10, VP2, VP4, ... | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-09-05 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K44
 
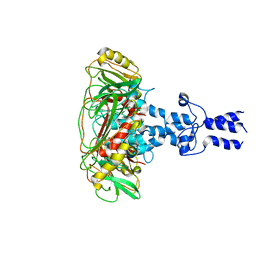 | | Structure of VP9 in Banna virus | | 分子名称: | VP9 | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-07-17 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K4A
 
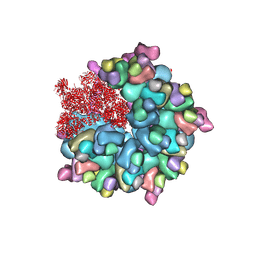 | | Structure of Banna virus core | | 分子名称: | VP10, VP2, VP8 | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-07-17 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K49
 
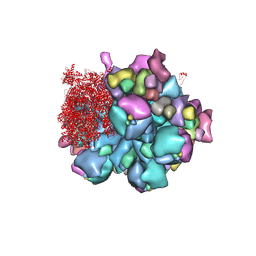 | | Structure of partial Banna virus | | 分子名称: | VP10, VP2, VP4, ... | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-07-17 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K43
 
 | |
8K42
 
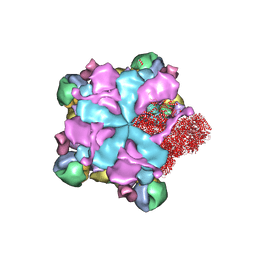 | | Structure of full Banna virus | | 分子名称: | VP10, VP2, VP4, ... | | 著者 | Li, Z, Cao, S. | | 登録日 | 2023-07-17 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
7Y6F
 
 | |
