4V2K
 
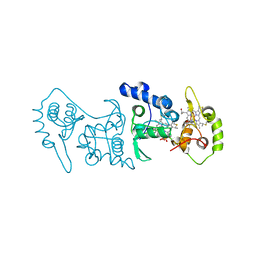 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | 分子名称: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | 著者 | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | 登録日 | 2014-10-10 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
4UWQ
 
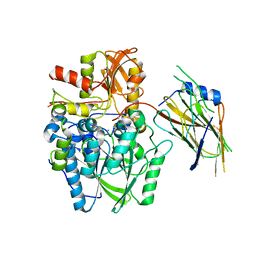 | | Crystal structure of the disulfide-linked complex of the thiosulfodyrolase SoxB with the carrier-protein SoxYZ from Thermus thermophilus | | 分子名称: | MANGANESE (II) ION, SOXY PROTEIN, SOXZ, ... | | 著者 | Grabarczyk, D.B, Chappell, P.E, Johnson, S, Stelzl, L.S, Lea, S.M, Berks, B.C. | | 登録日 | 2014-08-14 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7BII
 
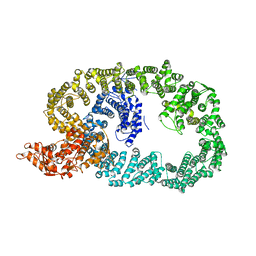 | | Crystal structure of Nematocida HUWE1 | | 分子名称: | E3 ubiquitin-protein ligase HUWE1 | | 著者 | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | 登録日 | 2021-01-12 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.037 Å) | | 主引用文献 | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
5OKC
 
 | |
6XWX
 
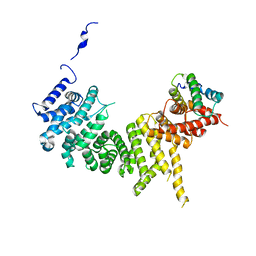 | | Crystal structure of the Tof1-Csm3 complex | | 分子名称: | Chromosome segregation in meiosis protein, Uncharacterized protein,Uncharacterized protein | | 著者 | Grabarczyk, D.B. | | 登録日 | 2020-01-24 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure and interactions of the Tof1-Csm3 (Timeless-Tipin) fork protection complex.
Nucleic Acids Res., 48, 2020
|
|
5OKI
 
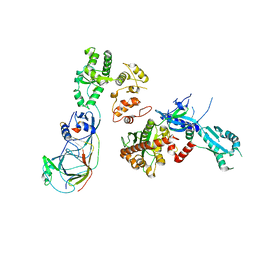 | |
6S1C
 
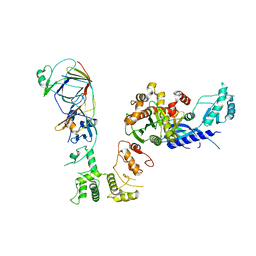 | | P3221 crystal form of the Ctf18-1-8/Pol2(1-528) complex | | 分子名称: | Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, DNA polymerase epsilon catalytic subunit A, ... | | 著者 | Grabarczyk, D.B. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (6.1 Å) | | 主引用文献 | Ctf18-RFC and DNA Pol ε form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication.
Nucleic Acids Res., 48, 2020
|
|
6S2F
 
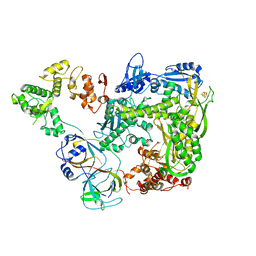 | |
6S2E
 
 | |
8ATX
 
 | |
8AUW
 
 | |
8ATU
 
 | |
8AUK
 
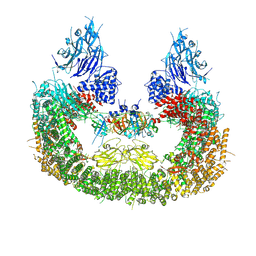 | | Cryo-EM structure of human BIRC6 in complex with HTRA2. | | 分子名称: | Baculoviral IAP repeat-containing protein 6, Serine protease HTRA2, mitochondrial, ... | | 著者 | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | 登録日 | 2022-08-25 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
6TUN
 
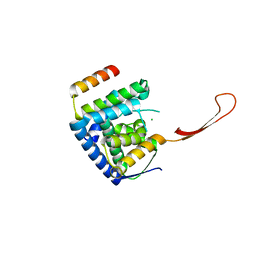 | | Helicase domain complex | | 分子名称: | CDK-activating kinase assembly factor MAT1, CHLORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPD | | 著者 | Sauer, F, Kisker, C. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair.
Nat Commun, 11, 2020
|
|
7NH3
 
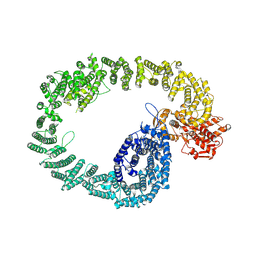 | | Nematocida Huwe1 in open conformation. | | 分子名称: | E3 ubiquitin-protein ligase HUWE1 | | 著者 | Petrova, O, Grishkovskaya, I, Grabarczyk, D.B, Kessler, D, Haselbach, D, Clausen, T. | | 登録日 | 2021-02-09 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (6.37 Å) | | 主引用文献 | Crystal structure of HUWE1: One ring to ubiquitinate them all
To Be Published
|
|
