4V7E
 
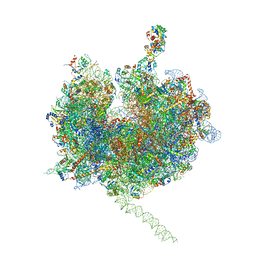 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | 著者 | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2013-11-22 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
4URD
 
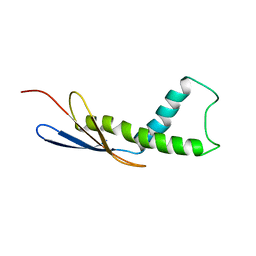 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | 分子名称: | TRIGGER FACTOR | | 著者 | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | 登録日 | 2014-06-27 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | 分子名称: | rRNA expansion segment ES27L in an "out" conformation | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-13 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V6M
 
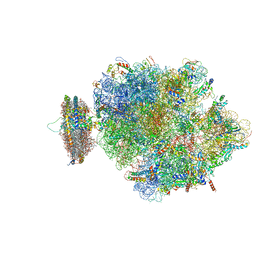 | | Structure of the ribosome-SecYE complex in the membrane environment | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | 著者 | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | 登録日 | 2011-02-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1TR8
 
 | |
7AFT
 
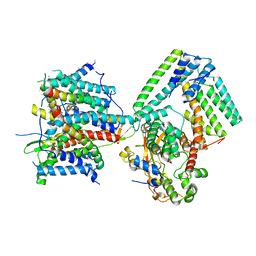 | | Cryo-EM structure of the signal sequence-engaged post-translational Sec translocon | | 分子名称: | Mating factor alpha-1,Mating factor alpha-1, Protein translocation protein SEC63, Protein transport protein SBH1, ... | | 著者 | Weng, T.-H, Beatrix, B, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | 登録日 | 2020-09-20 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the active post-translational Sec translocon.
Embo J., 40, 2021
|
|
8BQX
 
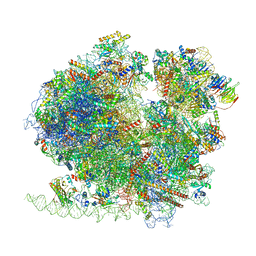 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
4CG5
 
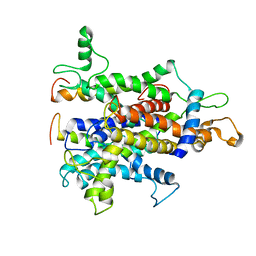 | | Cryo-EM of the Sec61-complex bound to the 80S ribosome translating a secretory substrate | | 分子名称: | PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT ALPHA ISOFORM 1, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT BETA, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT GAMMA | | 著者 | Gogala, M, Becker, T, Beatrix, B, Barrio-Garcia, C, Berninghausen, O, Beckmann, R. | | 登録日 | 2013-11-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion.
Nature, 506, 2014
|
|
8BQD
 
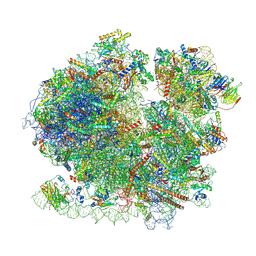 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BIP
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | 分子名称: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-02 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BJQ
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | 分子名称: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-05 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
4CG6
 
 | | Cryo-em of the Sec61-complex bound to the 80s ribosome translating a membrane-inserting substrate | | 分子名称: | PEPTIDE, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT ALPHA ISOFORM 1, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT BETA, ... | | 著者 | Gogala, M, Becker, T, Beatrix, B, Barrio-Garcia, C, Berninghausen, O, Beckmann, R. | | 登録日 | 2013-11-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion.
Nature, 506, 2014
|
|
4CG7
 
 | | Cryo-EM of the Sec61-complex bound to the idle 80S ribosome | | 分子名称: | PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT ALPHA ISOFORM 1, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT GAMMA, TRANSPORT PROTEIN SEC61 SUBUNIT BETA | | 著者 | Gogala, M, Becker, T, Beatrix, B, Barrio-Garcia, C, Berninghausen, O, Beckmann, R. | | 登録日 | 2013-11-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion.
Nature, 506, 2014
|
|
6R6G
 
 | | Structure of XBP1u-paused ribosome nascent chain complex with SRP. | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | 登録日 | 2019-03-27 | | 公開日 | 2019-07-10 | | 最終更新日 | 2019-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6R7Q
 
 | | Structure of XBP1u-paused ribosome nascent chain complex with Sec61. | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | 登録日 | 2019-03-29 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2018-12-16 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6ZZZ
 
 | |
6HD5
 
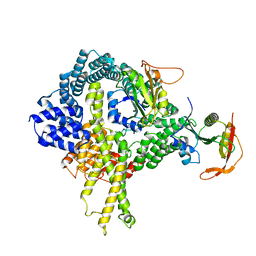 | | Cryo-EM structure of the ribosome-NatA complex | | 分子名称: | N-alpha-acetyltransferase NAT5, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1 | | 著者 | Knorr, A.G, Becker, T, Beckmann, R. | | 登録日 | 2018-08-17 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6HD7
 
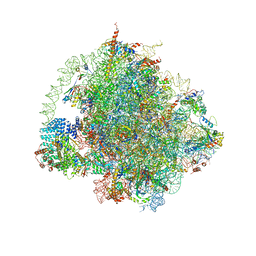 | | Cryo-EM structure of the ribosome-NatA complex | | 分子名称: | 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 5.8S rRNA, 5S rRNA, ... | | 著者 | Knorr, A.G, Becker, T, Beckmann, R. | | 登録日 | 2018-08-17 | | 公開日 | 2018-12-19 | | 最終更新日 | 2019-01-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
1B1A
 
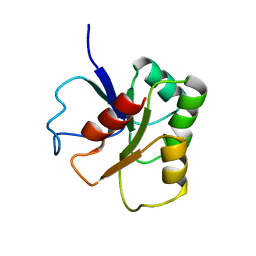 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | GLUTAMATE MUTASE | | 著者 | Hoffmann, B, Konrat, R, Bothe, H, Buckel, W, Kraeutler, B. | | 登録日 | 1998-11-19 | | 公開日 | 1999-07-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium.
Eur.J.Biochem., 263, 1999
|
|
6R6P
 
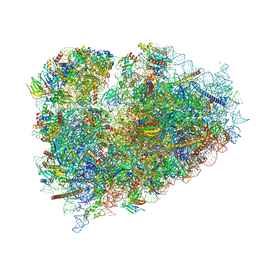 | | Structure of XBP1u-paused ribosome nascent chain complex (rotated state) | | 分子名称: | 18S rRNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | 登録日 | 2019-03-27 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6R5Q
 
 | | Structure of XBP1u-paused ribosome nascent chain complex (post-state) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | 登録日 | 2019-03-25 | | 公開日 | 2019-07-10 | | 最終更新日 | 2019-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
1I9C
 
 | |
1CB7
 
 | |
