7CKH
 
 | | Crystal structure of TMSiPheRS | | 分子名称: | Tyrosine--tRNA ligase | | 著者 | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | 登録日 | 2020-07-17 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79492676 Å) | | 主引用文献 | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
2LCC
 
 | |
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.293 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JWG
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-20 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
3L9R
 
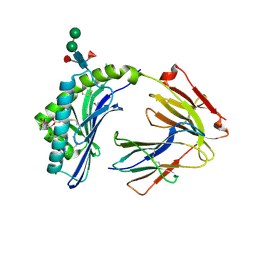 | | Crystal structure of bovine CD1b3 with endogenously bound ligands | | 分子名称: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zajonc, D.M, Girardi, E. | | 登録日 | 2010-01-05 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of bovine CD1b3 with endogenously bound ligands.
J.Immunol., 185, 2010
|
|
5ZVS
 
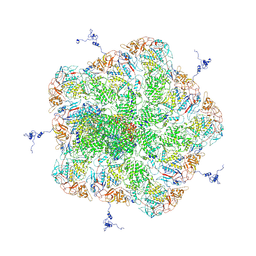 | |
7XPP
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | 著者 | Chen, Y.H, Wang, X.C, Zhang, C.R. | | 登録日 | 2022-05-05 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
3G11
 
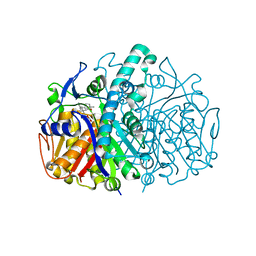 | | Structure of E. coli FabF(C163Q) in complex with dihydrophenyl platensimycin | | 分子名称: | 3-({3-[(1S,4S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxo-4-phenyldecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | 著者 | Soisson, S.M, Parthasarathy, G. | | 登録日 | 2009-01-29 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8GW7
 
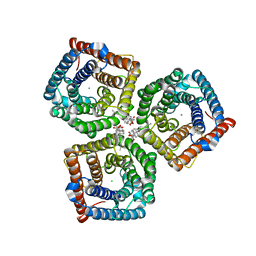 | | AtSLAC1 6D mutant in open state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2022-09-16 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW6
 
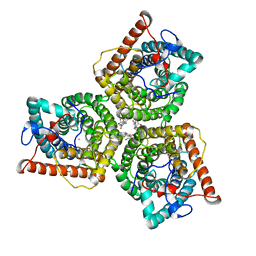 | | AtSLAC1 6D mutant in closed state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2022-09-16 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
3LQQ
 
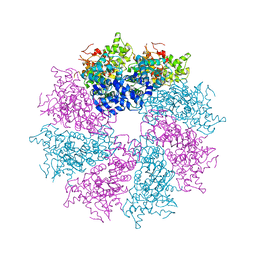 | | Structure of the CED-4 Apoptosome | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.534 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
8JV5
 
 | |
8JV6
 
 | |
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | 分子名称: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
3LRB
 
 | |
6AGJ
 
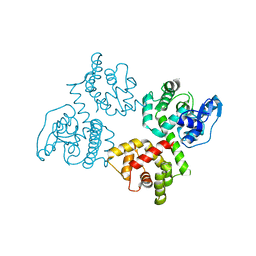 | |
5ZVT
 
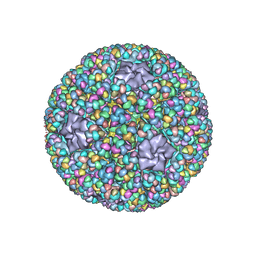 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | 分子名称: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | 著者 | Liu, H, Fang, Q, Cheng, L. | | 登録日 | 2018-05-12 | | 公開日 | 2018-07-04 | | 最終更新日 | 2018-07-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3G08
 
 | |
3LQR
 
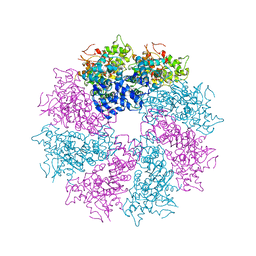 | | Structure of CED-4:CED-3 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.896 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
3G0Y
 
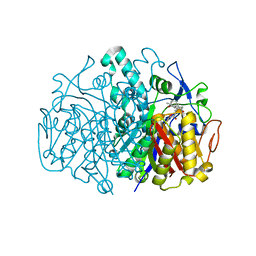 | | Structure of E. coli FabF(C163Q) in complex with dihydroplatensimycin | | 分子名称: | 3-({3-[(1S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxodecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | 著者 | Soisson, S.M, Parthasarathy, G. | | 登録日 | 2009-01-29 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5FKF
 
 | |
3LRC
 
 | |
3HPG
 
 | |
6AGI
 
 | | Crystal Structure of EFHA2 in Ca-binding State | | 分子名称: | CALCIUM ION, Calcium uptake protein 3, mitochondrial, ... | | 著者 | Yangfei, X, Xue, Y, Yuequan, S. | | 登録日 | 2018-08-11 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
