8JTA
 
 | |
8JTC
 
 | | Human VMAT2 complex with reserpine | | 分子名称: | Synaptic vesicular amine transporter, reserpine | | 著者 | Jiang, D.H, Wu, D. | | 登録日 | 2023-06-21 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JT9
 
 | | Human VMAT2 complex with ketanserin | | 分子名称: | 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quinazoline-2,4-dione, Synaptic vesicular amine transporter | | 著者 | Jiang, D.H, Wu, D. | | 登録日 | 2023-06-21 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wei, X, Zhang, Z. | | 登録日 | 2023-09-22 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | 分子名称: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | 著者 | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | 登録日 | 2015-07-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2017-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
5B7G
 
 | |
5B7Q
 
 | |
5B7N
 
 | |
5B7P
 
 | |
5VXA
 
 | | Structure of the human Mesh1-NADPH complex | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | 著者 | Rose, J, Zhou, P. | | 登録日 | 2017-05-23 | | 公開日 | 2018-05-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
1R9H
 
 | |
7KDT
 
 | |
6PIB
 
 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | 分子名称: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Cho, J, Zhou, P. | | 登録日 | 2019-06-26 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7TAE
 
 | |
5H3R
 
 | | Crystal Structure of mutant MarR C80S from E.coli complexed with operator DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*CP*TP*TP*GP*CP*CP*TP*GP*GP*GP*CP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*TP*AP*TP*TP*GP*CP*CP*CP*AP*GP*GP*CP*AP*AP*GP*TP*AP*T)-3'), Multiple antibiotic resistance protein MarR | | 著者 | Zhu, R, Lou, H, Hao, Z. | | 登録日 | 2016-10-26 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator.
J. Biol. Inorg. Chem., 22, 2017
|
|
8SAI
 
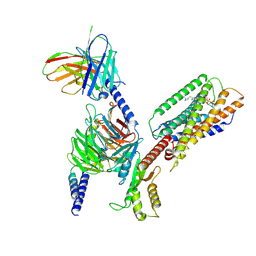 | | Cryo-EM structure of GPR34-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2023-04-01 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
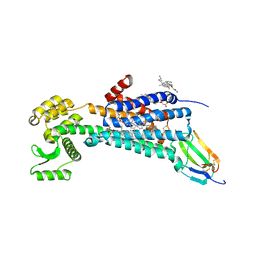
6WJC
 
 | |
7TAD
 
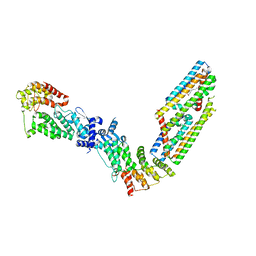 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | 分子名称: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | 著者 | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | 登録日 | 2021-12-20 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | 分子名称: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | 著者 | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | 登録日 | 2021-12-20 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8JGR
 
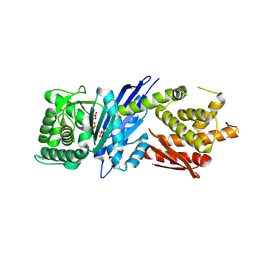 | |
8JGT
 
 | |
8JGW
 
 | |
8JGX
 
 | |
8JGU
 
 | |
