4R8T
 
 | | Structure of JEV protease | | 分子名称: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | 著者 | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | 登録日 | 2014-09-03 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.133 Å) | | 主引用文献 | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4YYX
 
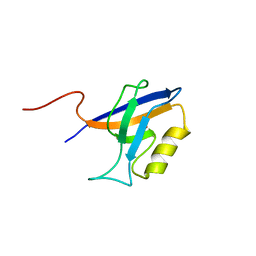 | |
4GUI
 
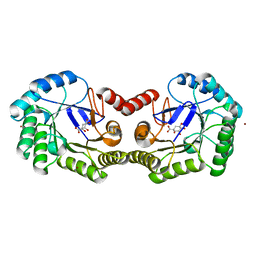 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | 著者 | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-29 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4GUJ
 
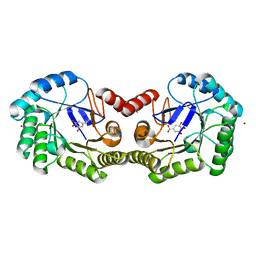 | | 1.50 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase, ZINC ION | | 著者 | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-29 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
3JS3
 
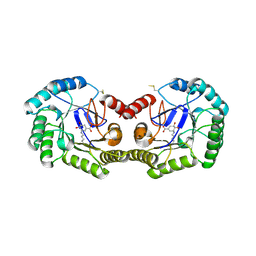 | | Crystal structure of type I 3-dehydroquinate dehydratase (aroD) from Clostridium difficile with covalent reaction intermediate | | 分子名称: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase | | 著者 | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-09-09 | | 公開日 | 2009-09-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
3TSV
 
 | |
3TSZ
 
 | |
3TSW
 
 | |
3OEX
 
 | | Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium with close loop conformation. | | 分子名称: | 3-dehydroquinate dehydratase, CHLORIDE ION | | 著者 | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-08-13 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.
Biochemistry, 50, 2011
|
|
1VMC
 
 | |
4OEP
 
 | |
4IUO
 
 | | 1.8 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) K170M Mutant in Complex with Quinate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase | | 著者 | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-21 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4OEO
 
 | |
5DND
 
 | |
5DNE
 
 | |
5DNC
 
 | |
