5XI2
 
 | | BRD4 bound with compound Bdi2 | | 分子名称: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Cao, D, Li, Y. | | 登録日 | 2017-04-25 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.909 Å) | | 主引用文献 | BRD4 bound with compound Bdi2
To Be Published
|
|
5XJ8
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the lysphosphatidic acid form | | 分子名称: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | 著者 | Li, Z, Tang, Y, Li, D. | | 登録日 | 2017-04-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XVU
 
 | | Crystal structure of the protein kinase CK2 catalytic domain from Plasmodium falciparum bound to ATP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | El Sahili, A, Lescar, J, Ruiz-Carrillo, D, Lin, J.Q. | | 登録日 | 2017-06-28 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The protein kinase CK2 catalytic domain from Plasmodium falciparum: crystal structure, tyrosine kinase activity and inhibition.
Sci Rep, 8, 2018
|
|
5XR1
 
 | |
8P2Z
 
 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | 分子名称: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | 登録日 | 2023-05-16 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P30
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | 分子名称: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | 登録日 | 2023-05-16 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P31
 
 | | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | | 分子名称: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | 登録日 | 2023-05-16 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
7TAS
 
 | |
7TAT
 
 | |
1ZUR
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1F) | | 分子名称: | CHLORIDE ION, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-4-phenyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | 著者 | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | 登録日 | 2005-05-31 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of spin labeled T4 Lysozyme (V131R1F)
To be Published
|
|
1ZS2
 
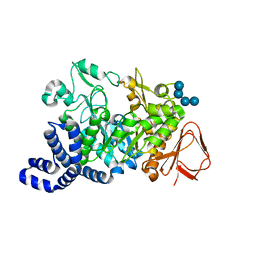 | | Amylosucrase Mutant E328Q in a ternary complex with sucrose and maltoheptaose | | 分子名称: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | 著者 | Skov, L.K, Mirza, O, Sprogoe, D, van der Veen, B.A, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | 登録日 | 2005-05-23 | | 公開日 | 2006-05-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of the Glu328Gln mutant of Neisseria polysaccharea amylosucrase in complex with sucrose and maltoheptaose
BIOCATAL.BIOTRANSFOR., 24, 2006
|
|
2ACU
 
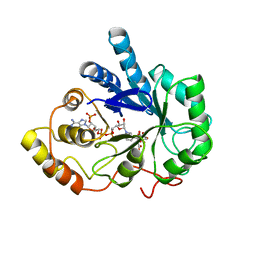 | | TYROSINE-48 IS THE PROTON DONOR AND HISTIDINE-110 DIRECTS SUBSTRATE STEREOCHEMICAL SELECTIVITY IN THE REDUCTION REACTION OF HUMAN ALDOSE REDUCTASE: ENZYME KINETICS AND THE CRYSTAL STRUCTURE OF THE Y48H MUTANT ENZYME | | 分子名称: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Bohren, K.M, Grimshaw, C.E, Lai, C.-J, Gabbay, K.H, Petsko, G.A, Harrison, D.H, Ringe, D. | | 登録日 | 1994-04-15 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme.
Biochemistry, 33, 1994
|
|
5AN7
 
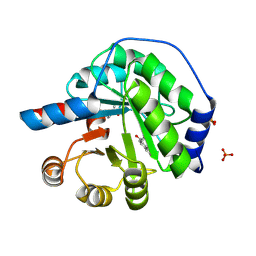 | | Structure of the engineered retro-aldolase RA95.5-8F with a bound 1,3-diketone inhibitor | | 分子名称: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, PHOSPHATE ION, RA95.5-8F | | 著者 | Obexer, R, Mittl, P.R.E, Hilvert, D. | | 登録日 | 2015-09-04 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
2A86
 
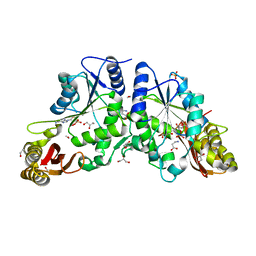 | | Crystal structure of A Pantothenate synthetase complexed with AMP and beta-alanine | | 分子名称: | ADENOSINE MONOPHOSPHATE, BETA-ALANINE, ETHANOL, ... | | 著者 | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2005-07-07 | | 公開日 | 2006-02-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
208D
 
 | | HIGH-RESOLUTION STRUCTURE OF A DNA HELIX FORMING (C.G)*G BASE TRIPLETS | | 分子名称: | DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION | | 著者 | Van Meervelt, L, Vlieghe, D, Dautant, A, Gallois, B, Precigoux, G, Kennard, O. | | 登録日 | 1995-04-26 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | High-resolution structure of a DNA helix forming (C.G)*G base triplets.
Nature, 374, 1995
|
|
5J6Q
 
 | | Cwp8 from Clostridium difficile | | 分子名称: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | 著者 | Renko, M, Usenik, A, Turk, D. | | 登録日 | 2016-04-05 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
2A84
 
 | | Crystal structure of A Pantothenate synthetase complexed with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2005-07-07 | | 公開日 | 2006-02-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A88
 
 | | Crystal structure of A Pantothenate synthetase, apo enzyme in C2 space group | | 分子名称: | GLYCEROL, Pantoate--beta-alanine ligase, SULFATE ION | | 著者 | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2005-07-07 | | 公開日 | 2006-02-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
5AOU
 
 | | Structure of the engineered retro-aldolase RA95.5-8F apo | | 分子名称: | 1,2-ETHANEDIOL, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, PHOSPHATE ION | | 著者 | Obexer, R, Mittl, P, Hilvert, D. | | 登録日 | 2015-09-11 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
2A7X
 
 | | Crystal Structure of A Pantothenate synthetase complexed with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Pantoate-beta-alanine ligase, ... | | 著者 | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2005-07-06 | | 公開日 | 2006-02-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
1ZWN
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1B) | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | 著者 | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | 登録日 | 2005-06-03 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of spin labeled T4 Lysozyme (V131R1B)
To be Published
|
|
2A8H
 
 | | Crystal structure of catalytic domain of TACE with Thiomorpholine Sulfonamide Hydroxamate inhibitor | | 分子名称: | 4-({4-[(4-AMINOBUT-2-YNYL)OXY]PHENYL}SULFONYL)-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | 著者 | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Jin, G, Barone, D, Skotnicki, J.S. | | 登録日 | 2005-07-08 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Acetylenic TACE inhibitors. Part 3: Thiomorpholine sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2A4T
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R7) | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | 著者 | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | 登録日 | 2005-06-29 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of spin labeled T4 Lysozyme (V131R7
To be Published
|
|
5AP8
 
 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | 分子名称: | TSR3 | | 著者 | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | 登録日 | 2015-09-14 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.246 Å) | | 主引用文献 | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
2K9A
 
 | | The Solution Structure of the Arl2 Effector, BART | | 分子名称: | ADP-ribosylation factor-like protein 2-binding protein | | 著者 | Bailey, L.K, Campbell, L.J, Evetts, K.A, Littlefield, K, Rajendra, E, Nietlispach, D, Owen, D, Mott, H.R. | | 登録日 | 2008-10-06 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of Binder of Arl2 (BART) Reveals a Novel G Protein Binding Domain: IMPLICATIONS FOR FUNCTION.
J.Biol.Chem., 284, 2009
|
|
