8HNC
 
 | | Cryo-EM structure of human OATP1B1 in complex with bilirubin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | 著者 | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HND
 
 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | 著者 | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
7SA3
 
 | | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | 著者 | Gisriel, C.J, Bryant, D.A, Brudvig, G.W. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.25 Å) | | 主引用文献 | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
J.Biol.Chem., 298, 2021
|
|
6VIK
 
 | |
6ULY
 
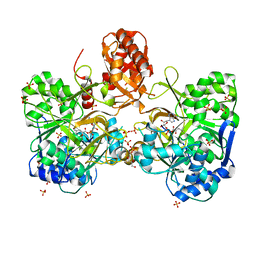 | | Adenylation domain of a keto acid-selecting NRPS module bound to keto acyl adenylate space group P212121 | | 分子名称: | 5'-O-{(S)-hydroxy[(4-methyl-2-oxopentanoyl)oxy]phosphoryl}adenosine, Amino acid adenylation domain-containing protein, GLYCEROL, ... | | 著者 | Alonzo, D.A, Chiche-Lapierre, C, Schmeing, T.M. | | 登録日 | 2019-10-08 | | 公開日 | 2020-02-19 | | 最終更新日 | 2020-05-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis.
Nat.Chem.Biol., 16, 2020
|
|
5FDL
 
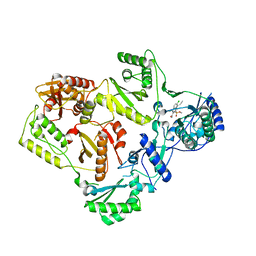 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | 分子名称: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | 著者 | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | 登録日 | 2015-12-16 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
1HVU
 
 | |
7DRS
 
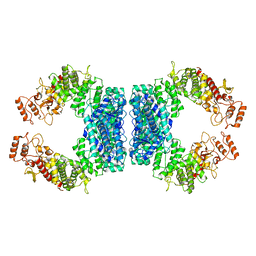 | | Structure of SspE_40224 | | 分子名称: | SspE protein | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
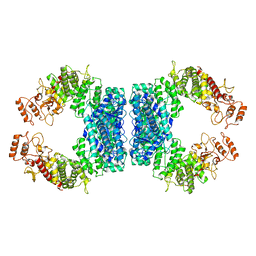 | | Structure of SspE-R100A protein | | 分子名称: | SspE protein | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.48 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7S5S
 
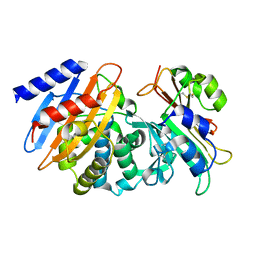 | | CTX-M-15 WT in complex with BLIP WT | | 分子名称: | Beta-lactamase, Beta-lactamase inhibitory protein | | 著者 | Lu, S, Palzkill, T, Hu, L.Y, Prasad, B.V.V, Sankaran, B. | | 登録日 | 2021-09-11 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | An active site loop toggles between conformations to control antibiotic hydrolysis and inhibition potency for CTX-M beta-lactamase drug-resistance enzymes.
Nat Commun, 13, 2022
|
|
6AX1
 
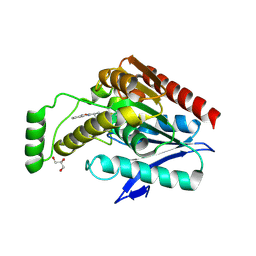 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | 分子名称: | 1,1,1,3,3,3-hexafluoropropan-2-yl 3-(3-phenyl-1,2,4-oxadiazol-5-yl)azetidine-1-carboxylate, GLYCEROL, Monoglyceride lipase | | 著者 | Pandit, J. | | 登録日 | 2017-09-06 | | 公開日 | 2017-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase.
J. Med. Chem., 60, 2017
|
|
7EO0
 
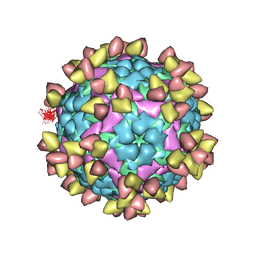 | |
4HCP
 
 | |
3UDC
 
 | | Crystal structure of a membrane protein | | 分子名称: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | 著者 | Li, W, Ge, J, Yang, M. | | 登録日 | 2011-10-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.355 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WY1
 
 | | Crystal structure of human BACE-1 bound to Compound 24B | | 分子名称: | (4aR,8aS)-8a-(2,4-difluorophenyl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1 | | 著者 | Vajdos, F.F, Parris, K. | | 登録日 | 2014-11-14 | | 公開日 | 2015-03-04 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of a Series of Efficient, Centrally Efficacious BACE1 Inhibitors through Structure-Based Drug Design.
J.Med.Chem., 58, 2015
|
|
4Q9S
 
 | | Crystal Structure of human Focal Adhesion Kinase (Fak) bound to Compound1 (3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | 分子名称: | 3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, Focal adhesion kinase 1 | | 著者 | Argiriadi, M.A, George, D.M. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
4Q9Z
 
 | | Human Protein Kinase C Theta in Complex with Compound35 ((1R)-9-(AZETIDIN-3-YLAMINO)-1,8-DIMETHYL-3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | 分子名称: | (1R)-9-(azetidin-3-ylamino)-1,8-dimethyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, HUMAN PROTEIN KINASE C THETA, SODIUM ION | | 著者 | Argiriadi, M.A, George, D.M. | | 登録日 | 2014-05-02 | | 公開日 | 2014-07-02 | | 最終更新日 | 2015-01-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
4M49
 
 | |
8HVU
 
 | |
8HVV
 
 | |
8HVX
 
 | |
4Y2D
 
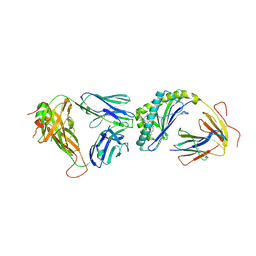 | | Crystal structure of the mCD1d/7DW8-5/iNKTCR ternary complex | | 分子名称: | 11-(4-fluorophenyl)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]undecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zajonc, D.M, Yu, E.D. | | 登録日 | 2015-02-09 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural modifications of alphaGalCer in both lipid and carbohydrate moiety influence activation of murine and human iNKT cells
To Be Published
|
|
8HVZ
 
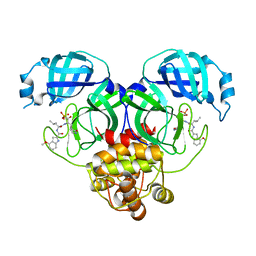 | |
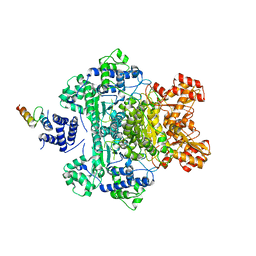
4QOY
 
 | |
8HVW
 
 | |
