5OSG
 
 | | Structure of KSRP in context of Leishmania donovani 80S | | 分子名称: | 18S rRNA, 40S ribosomal protein S6, RNA binding protein, ... | | 著者 | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | 登録日 | 2017-08-17 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
3K3O
 
 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | 著者 | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | 登録日 | 2009-10-03 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
2MQG
 
 | |
3K3N
 
 | | Crystal structure of the catalytic core domain of human PHF8 | | 分子名称: | FE (II) ION, PHD finger protein 8 | | 著者 | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | 登録日 | 2009-10-03 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
5H05
 
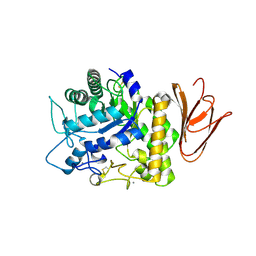 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | 分子名称: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | He, C, Liu, Y. | | 登録日 | 2016-10-03 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
3K1Q
 
 | | Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics | | 分子名称: | Core protein VP6, Outer capsid VP5, Outer capsid VP7, ... | | 著者 | Cheng, L.P, Zhu, J, Hiu, W.H, Zhang, X.K, Honig, B, Fang, Q, Zhou, Z.H. | | 登録日 | 2009-09-28 | | 公開日 | 2010-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Backbone Model of an Aquareovirus Virion by Cryo-Electron Microscopy and Bioinformatics
J.Mol.Biol., 397, 2010
|
|
1O3R
 
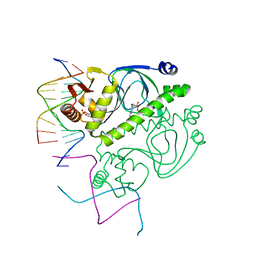 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
4M5F
 
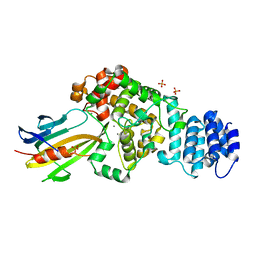 | | complex structure of Tse3-Tsi3 | | 分子名称: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | 著者 | Gu, L.C. | | 登録日 | 2013-08-08 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
8K3C
 
 | |
5NSR
 
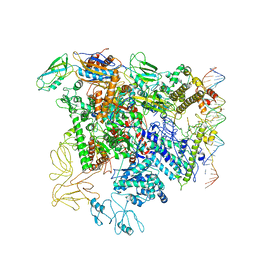 | | Cryo-EM structure of RNA polymerase-sigma54 holo enzyme with promoter DNA closed complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | 登録日 | 2017-04-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
1PFN
 
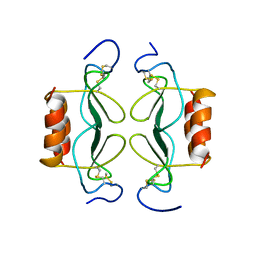 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | 分子名称: | PF4-M2 CHIMERA | | 著者 | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | 登録日 | 1995-07-18 | | 公開日 | 1996-01-29 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFM
 
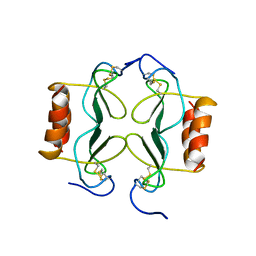 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | 分子名称: | PF4-M2 CHIMERA | | 著者 | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | 登録日 | 1995-07-18 | | 公開日 | 1996-01-29 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
8P5Y
 
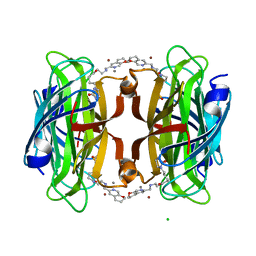 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | 分子名称: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Lau, K, Wang, W, Pojer, F, Larabi, A. | | 登録日 | 2023-05-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Z
 
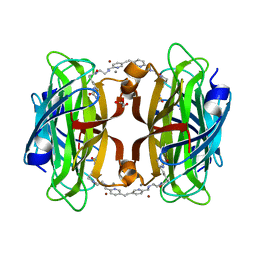 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | 分子名称: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | 著者 | Lau, K, Wang, W, Pojer, F, Larabi, A. | | 登録日 | 2023-05-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1O3Q
 
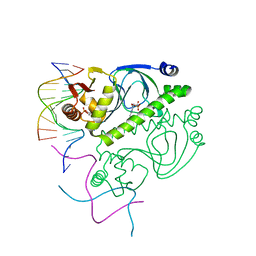 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3T
 
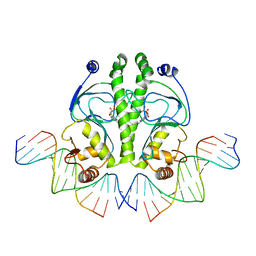 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex:
DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
5BQX
 
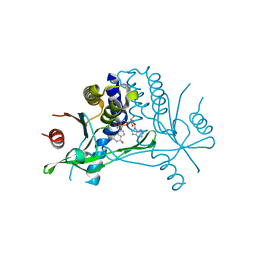 | | Crystal structure of human STING in complex with 3'2'-cGAMP | | 分子名称: | 3'2'-cGAMP, Stimulator of interferon genes protein | | 著者 | Wu, J, Zhang, X, Chen, Z.J, Chen, C. | | 登録日 | 2015-05-29 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2H2I
 
 | | The Structural basis of Sirtuin Substrate Affinity | | 分子名称: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | 著者 | Cosgrove, M.S, Wolberger, C. | | 登録日 | 2006-05-18 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
2H2G
 
 | |
2H2D
 
 | |
2H2F
 
 | |
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
5H7P
 
 | |
