1AQL
 
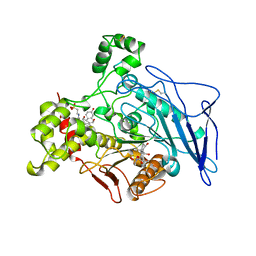 | |
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
2L88
 
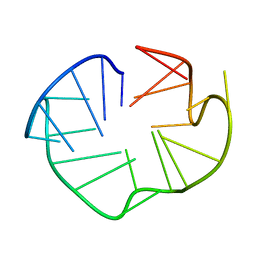 | |
2QRV
 
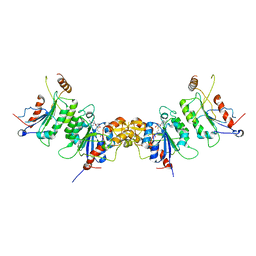 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Jia, D, Cheng, X. | | 登録日 | 2007-07-29 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
6K5H
 
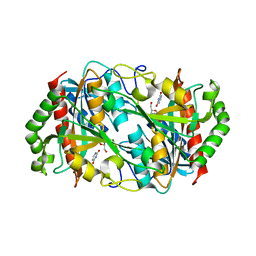 | |
8DDI
 
 | |
8DDM
 
 | |
5ZUH
 
 | |
4Q9V
 
 | | Crystal structure of TIPE3 | | 分子名称: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | 著者 | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | 登録日 | 2014-05-02 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
6EAY
 
 | |
5XJY
 
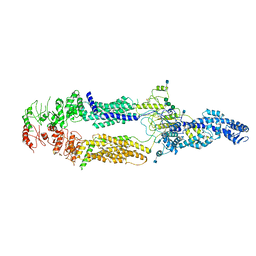 | | Cryo-EM structure of human ABCA1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette sub-family A member 1, ... | | 著者 | Qian, H.W, Yan, N, Gong, X. | | 登録日 | 2017-05-04 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the Human Lipid Exporter ABCA1.
Cell, 169, 2017
|
|
3MO2
 
 | | human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E67 | | 分子名称: | 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | 著者 | Chang, Y, Horton, J.R, Cheng, X. | | 登録日 | 2010-04-22 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
1U6Q
 
 | | Substituted 2-Naphthamadine inhibitors of Urokinase | | 分子名称: | TRANS-6-(2-PHENYLCYCLOPROPYL)-NAPHTHALENE-2-CARBOXAMIDINE, Urokinase-type plasminogen activator | | 著者 | Bruncko, M, McClellan, W, Wendt, M.D, Sauer, D.R, Geyer, A, Dalton, C.R, Kaminski, M.K, Nienaber, V.L, Rockway, T.R, Giranda, V.L. | | 登録日 | 2004-07-30 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Naphthamidine urokinase plasminogen activator inhibitors with improved pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | 分子名称: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6YE2
 
 | |
6YE1
 
 | | Human Ecto-5'-nucleotidase (CD73) in complex with the AMPCP derivative A894 (compound 2n in publication) in the closed form (crystal form IV) | | 分子名称: | 5'-nucleotidase, ZINC ION, [(2~{R},3~{S},4~{R},5~{R})-5-[2-chloranyl-6-(cyclopentylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxymethylphosphonic acid | | 著者 | Scaletti, E, Strater, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Discovery of Potent and Selective Methylenephosphonic Acid CD73 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8GUE
 
 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | 著者 | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | 登録日 | 2022-09-11 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
5GNF
 
 | |
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2014-04-09 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
3MO0
 
 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E11 | | 分子名称: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | 著者 | Chang, Y, Horton, J.R, Cheng, X. | | 登録日 | 2010-04-22 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
5EDS
 
 | | Crystal structure of human PI3K-gamma in complex with benzimidazole inhibitor 5 | | 分子名称: | 4-azanyl-6-[[(1~{S})-1-[6-fluoranyl-1-(3-methylsulfonylphenyl)benzimidazol-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2015-10-21 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery, Optimization, and in Vivo Evaluation of Benzimidazole Derivatives AM-8508 and AM-9635 as Potent and Selective PI3K delta Inhibitors.
J.Med.Chem., 59, 2016
|
|
7L7D
 
 | |
7L7E
 
 | |
2HNA
 
 | | Solution Structure of a bacterial apo-flavodoxin | | 分子名称: | Protein mioC | | 著者 | Hu, Y, Jin, C. | | 登録日 | 2006-07-12 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
