8D9N
 
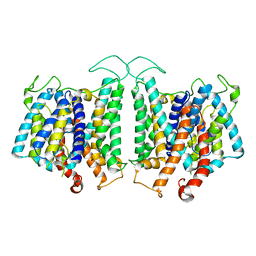 | | CryoEM structures of bAE1 captured in multiple states. | | 分子名称: | Anion exchange protein | | 著者 | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | 登録日 | 2022-06-10 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8E34
 
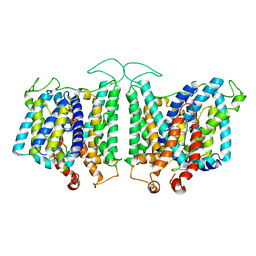 | | CryoEM structures of bAE1 captured in multiple states | | 分子名称: | Anion exchange protein | | 著者 | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | 登録日 | 2022-08-16 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8EEQ
 
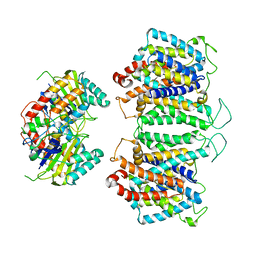 | | CryoEM structures of bAE1 captured in multiple states. | | 分子名称: | Anion exchange protein | | 著者 | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | 登録日 | 2022-09-07 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
4ZMJ
 
 | | Crystal Structure of Ligand-Free BG505 SOSIP.664 HIV-1 Env Trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | 著者 | Kwon, Y.D, Kwong, P.D. | | 登録日 | 2015-05-04 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5C77
 
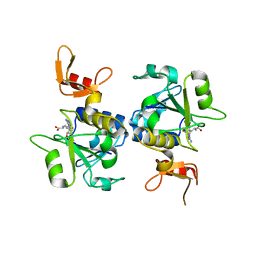 | | A novel protein arginine methyltransferase | | 分子名称: | Protein arginine N-methyltransferase SFM1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Lv, F, Zhang, T, Ding, J. | | 登録日 | 2015-06-24 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
5C74
 
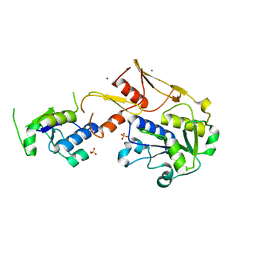 | |
4NZW
 
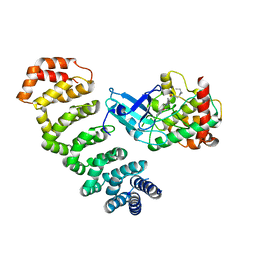 | | Crystal Structure of STK25-MO25 Complex | | 分子名称: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | 著者 | Feng, M, Hao, Q, Zhou, Z.C. | | 登録日 | 2013-12-13 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.583 Å) | | 主引用文献 | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
4O27
 
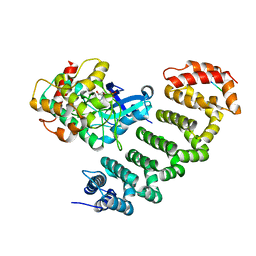 | | Crystal structure of MST3-MO25 complex with WIF motif | | 分子名称: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | 著者 | Hao, Q, Feng, M, Zhou, Z.C. | | 登録日 | 2013-12-16 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.185 Å) | | 主引用文献 | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
4FI9
 
 | | Structure of human SUN-KASH complex | | 分子名称: | Nesprin-2, SUN domain-containing protein 2 | | 著者 | Wang, W.J, Shi, Z.B. | | 登録日 | 2012-06-08 | | 公開日 | 2012-07-18 | | 最終更新日 | 2013-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
6UTK
 
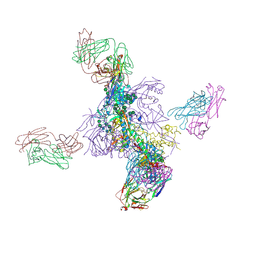 | |
2KBE
 
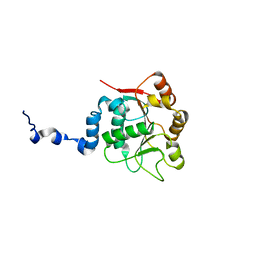 | |
2KBF
 
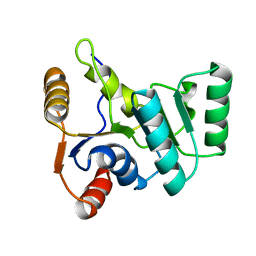 | |
1HDK
 
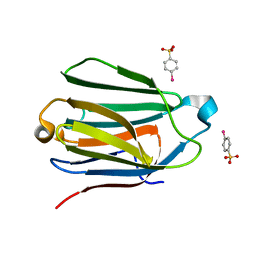 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | 分子名称: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | 著者 | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | 登録日 | 2000-11-16 | | 公開日 | 2001-11-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
1G86
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN/N-ETHYLMALEIMIDE COMPLEX | | 分子名称: | CHARCOT-LEYDEN CRYSTAL PROTEIN, N-ETHYLMALEIMIDE | | 著者 | Ackerman, S.J, Liu, L, Kwatia, M.A, Savage, M.P, Leonidas, D.D, Swaminathan, G.J, Acharya, K.R. | | 登録日 | 2000-11-16 | | 公開日 | 2002-06-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion.
J.Biol.Chem., 277, 2002
|
|
4DF3
 
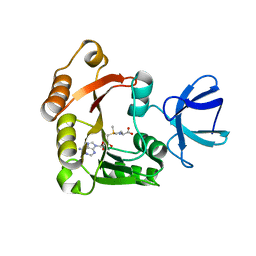 | |
2HNK
 
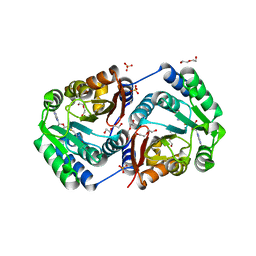 | | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans | | 分子名称: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent O-methyltransferase, ... | | 著者 | Hou, X, Wei, Z, Gong, W. | | 登録日 | 2006-07-13 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans.
J.Struct.Biol., 159, 2007
|
|
4QEI
 
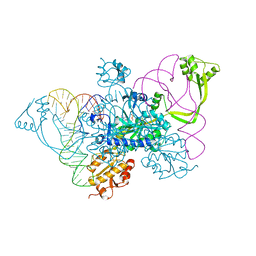 | | Two distinct conformational states of GlyRS captured in crystal lattice | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glycine--tRNA ligase, tRNA-Gly-CCC-2-2 | | 著者 | Xie, W, Qin, X, Deng, X, Zhang, Q, Li, Q. | | 登録日 | 2014-05-16 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.875 Å) | | 主引用文献 | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
5XY9
 
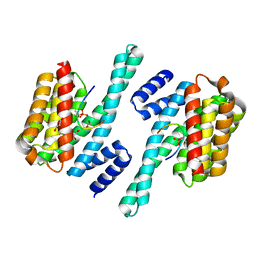 | | Structure of the MST4 and 14-3-3 complex | | 分子名称: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2017-07-06 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
6WZU
 
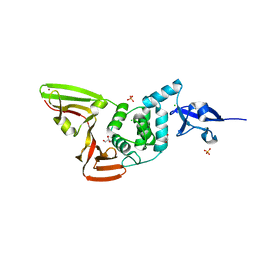 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-14 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6WRH
 
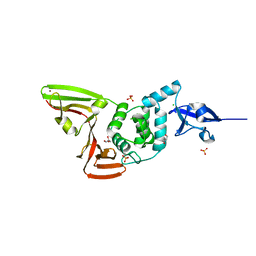 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6XG3
 
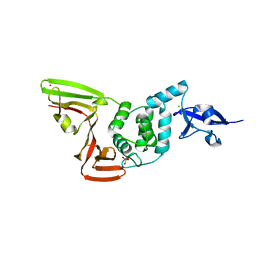 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | 分子名称: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
8XRY
 
 | |
8XNG
 
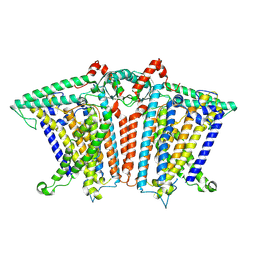 | |
8XW3
 
 | |
8XS4
 
 | |
