1C7P
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME WITH FOUR EXTRA RESIDUES (EAEA) AT THE N-TERMINAL | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Goda, S, Takano, K, Yamagata, Y, Katakura, Y, Yutani, K. | | 登録日 | 2000-02-29 | | 公開日 | 2000-04-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Effect of extra N-terminal residues on the stability and folding of human lysozyme expressed in Pichia pastoris.
Protein Eng., 13, 2000
|
|
1DI5
 
 | |
1DI3
 
 | |
1DI4
 
 | |
7YVW
 
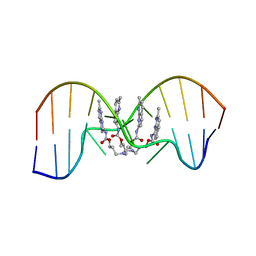 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | 分子名称: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | 著者 | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | 登録日 | 2022-08-19 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
6QIS
 
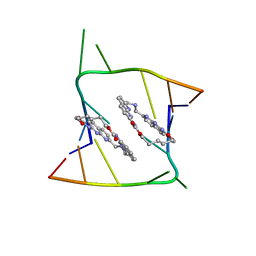 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model II) | | 分子名称: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3'), SULFATE ION | | 著者 | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | 登録日 | 2019-01-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIV
 
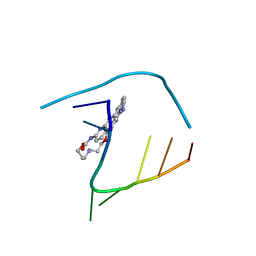 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL4 compound | | 分子名称: | CMBL4, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | 著者 | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | 登録日 | 2019-01-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIQ
 
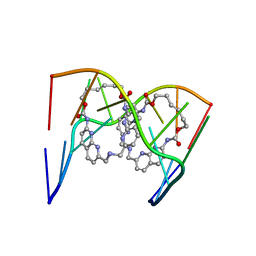 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL3a compound | | 分子名称: | CMBL3a, RNA (5'-R(*GP*CP*AP*G)-D(P*(CSL))-R(P*AP*GP*C)-3') | | 著者 | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Micura, R, Nakatani, K. | | 登録日 | 2019-01-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.519 Å) | | 主引用文献 | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIT
 
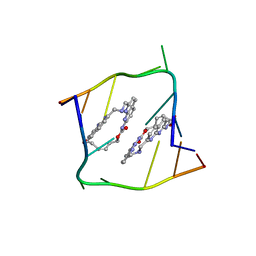 | | Crystal structure of CAG repeats with synthetic CMBL3b compound | | 分子名称: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | 著者 | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | 登録日 | 2019-01-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
1JJU
 
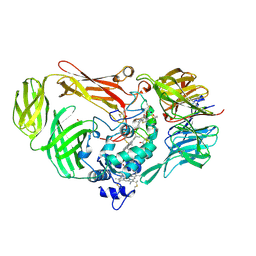 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | 著者 | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | 登録日 | 2001-07-09 | | 公開日 | 2001-12-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6KJL
 
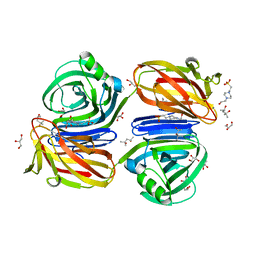 | | Xylanase J from Bacillus sp. strain 41M-1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | 登録日 | 2019-07-22 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
7WSS
 
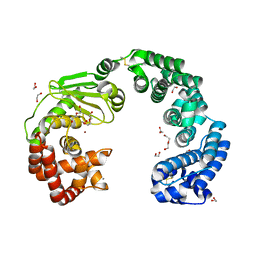 | | Collagenase from Grimontia (Vibrio) hollisae 1706B | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | 登録日 | 2022-02-01 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
7WAG
 
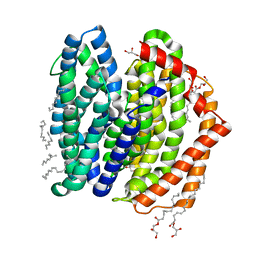 | | Crystal structure of MurJ squeezed form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipid II flippase MurJ | | 著者 | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | 登録日 | 2021-12-14 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAX
 
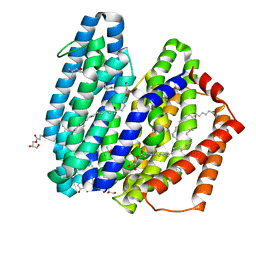 | | MurJ inward occluded form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, lipid II flippase MurJ | | 著者 | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | 登録日 | 2021-12-15 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAW
 
 | | MurJ inward closed form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, lipid II flippase MurJ | | 著者 | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | 登録日 | 2021-12-15 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
8X3H
 
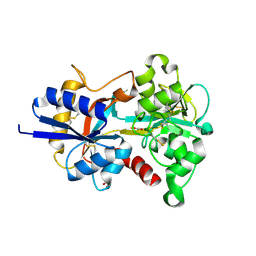 | |
7XEB
 
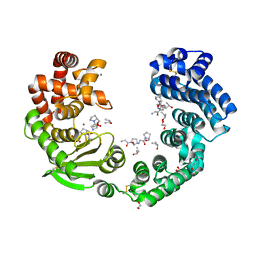 | | Collagenase from Grimontia (Vibrio) hollisae 1706B complexed with Gly-Pro-Hyp | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLY-PRO-HYP peptide, ... | | 著者 | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | 登録日 | 2022-03-30 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | 分子名称: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | 著者 | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | 登録日 | 2019-11-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6IZP
 
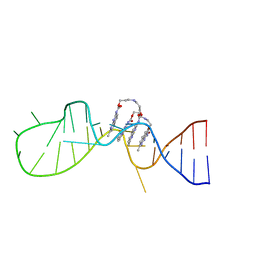 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | 分子名称: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | 著者 | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
5ZFS
 
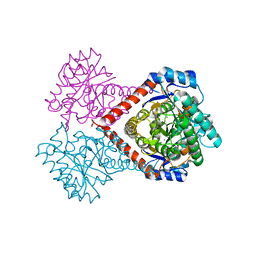 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | 分子名称: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | 登録日 | 2018-03-07 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1X12
 
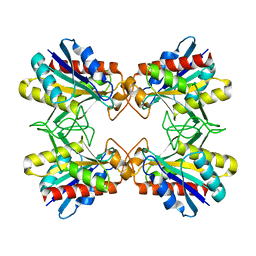 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192D) from a Hyperthermophile, Pyrococcus furiosus | | 分子名称: | Pyrrolidone-carboxylate peptidase | | 著者 | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | 登録日 | 2005-03-31 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1X10
 
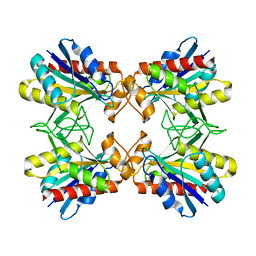 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192A) from a Hyperthermophile, Pyrococcus furiosus | | 分子名称: | Pyrrolidone-carboxylate peptidase | | 著者 | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | 登録日 | 2005-03-31 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
6QIR
 
 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model I) | | 分子名称: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | 著者 | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | 登録日 | 2019-01-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.531 Å) | | 主引用文献 | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
5ZEA
 
 | | Crystal structure of the nucleotide-free mutant A3B3 | | 分子名称: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | 著者 | Maruyama, S, Suzuki, K, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | 登録日 | 2018-02-27 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.384 Å) | | 主引用文献 | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Suzuki, M, Takita, T, Nakatani, K. | | 登録日 | 2019-07-24 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
