7N26
 
 | |
6UYF
 
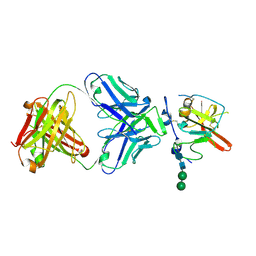 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v1 redesigned core from genotype 6a bound to broadly neutralizing antibody AR3B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Zhu, J. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6UYM
 
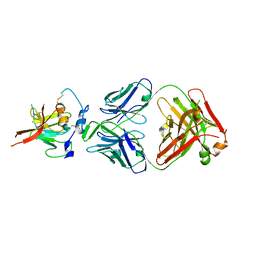 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v6 redesigned core from genotype 1a bound to broadly neutralizing antibody AR3C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Zhu, J. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.848 Å) | | 主引用文献 | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6UYG
 
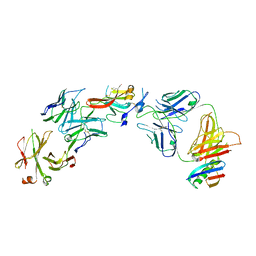 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2c3 core from genotype 6a bound to broadly neutralizing antibody AR3A and non neutralizing antibody E1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Zhu, J. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.375 Å) | | 主引用文献 | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
7N21
 
 | | NMR structure of AnIB-OH | | 分子名称: | Alpha-conotoxin AnIB | | 著者 | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N23
 
 | |
7N22
 
 | |
7N0T
 
 | |
7N20
 
 | | NMR structure of native AnIB | | 分子名称: | Alpha-conotoxin AnIB | | 著者 | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
6V98
 
 | | Crystal structure of Type VI secretion system effector, TseH (VCA0285) | | 分子名称: | CALCIUM ION, Cysteine hydrolase | | 著者 | Watanabe, N, Hersch, S.J, Dong, T.G, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2020-05-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Envelope stress responses defend against type six secretion system attacks independently of immunity proteins.
Nat Microbiol, 5, 2020
|
|
6X2W
 
 | |
6X2U
 
 | |
4YND
 
 | | The Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2 | | 分子名称: | N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[(2R)-2-hydroxy-2-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | 著者 | Sweis, R.F, Wang, Z, Algire, M, Arrowsmith, C.H, Brown, P.J, Chiang, G.C, Guo, J, Jakob, C.G, Kennedy, S, Li, F, Soni, N.B, Vedadi, M, Pappano, W.N. | | 登録日 | 2015-03-09 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2.
Acs Med.Chem.Lett., 6, 2015
|
|
6HF5
 
 | | Crystal Structure of the Acquired VIM-2 Metallo-beta-Lactamase in Complex with ANT-431 Inhibitor | | 分子名称: | 5-(pyridin-3-ylsulfonylamino)-1,3-thiazole-4-carboxylic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | 著者 | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | 登録日 | 2018-08-21 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | SAR Studies Leading to the Identification of a Novel Series of Metallo-beta-lactamase Inhibitors for the Treatment of Carbapenem-Resistant Enterobacteriaceae Infections That Display Efficacy in an Animal Infection Model.
Acs Infect Dis., 5, 2019
|
|
6HAZ
 
 | | Crystal structure of the bromodomain of human SMARCA2 in complex with SMARCA-BD ligand | | 分子名称: | 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Probable global transcription activator SNF2L2, ZINC ION | | 著者 | Bader, G, Steurer, S, Weiss-Puxbaum, A, Zoephel, A, Roy, M, Ciulli, A. | | 登録日 | 2018-08-09 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6HAX
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | 分子名称: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | 登録日 | 2018-08-09 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6HR2
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | 分子名称: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | 著者 | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | 登録日 | 2018-09-26 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6HAY
 
 | | Crystal structure of PROTAC 1 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | 分子名称: | (2~{S},4~{R})-~{N}-[[2-[2-[2-[2-[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]ethoxy]ethoxy]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | 登録日 | 2018-08-09 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
400D
 
 | |
5IA7
 
 | |
6X2P
 
 | | Crystal Structure of the Mek1NES peptide bound to CRM1 | | 分子名称: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | 著者 | Baumhardt, J.M. | | 登録日 | 2020-05-20 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2V
 
 | |
6X2O
 
 | |
6X2X
 
 | |
6X2R
 
 | |
