6LOL
 
 | | The crystal structure of full length IpaH9.8 | | 分子名称: | E3 ubiquitin-protein ligase ipaH9.8 | | 著者 | Ye, Y, Huang, H. | | 登録日 | 2020-01-06 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
5VT7
 
 | | ABA-mimicking ligand AMC1beta in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | 1-(3-chloro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | 著者 | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | 登録日 | 2017-05-15 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.624 Å) | | 主引用文献 | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
3QNQ
 
 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | 著者 | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2011-02-08 | | 公開日 | 2011-04-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.295 Å) | | 主引用文献 | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
3PJZ
 
 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | 分子名称: | POTASSIUM ION, Potassium uptake protein TrkH | | 著者 | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2010-11-10 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.506 Å) | | 主引用文献 | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
3OF7
 
 | | The Crystal Structure of Prp20p from Saccharomyces cerevisiae and Its Binding Properties to Gsp1p and Histones | | 分子名称: | Regulator of chromosome condensation | | 著者 | Wu, F, Liu, Y, Zhu, Z, Huang, H, Ding, B, Wu, J, Shi, Y. | | 登録日 | 2010-08-14 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The 1.9A crystal structure of Prp20p from Saccharomyces cerevisiae and its binding properties to Gsp1p and histones.
J.Struct.Biol., 174, 2011
|
|
7VKA
 
 | | Crystal Structure of GH3.6 in complex with an inhibitor | | 分子名称: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | 著者 | Wang, N, Luo, M, Bao, H, Huang, H. | | 登録日 | 2021-09-29 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4IIO
 
 | | Crystal Structure of the Second SH3 Domain of ITSN2 Bound with a Synthetic Peptide | | 分子名称: | CHLORIDE ION, Intersectin-2, SULFATE ION, ... | | 著者 | Dong, A, Guan, X, Huang, H, Gu, J, Tempel, W, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2012-12-20 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Second SH3 Domain of ITSN2 Bound with a Synthetic Peptide
TO BE PUBLISHED
|
|
4IIM
 
 | | Crystal structure of the Second SH3 Domain of ITSN1 bound with a synthetic peptide | | 分子名称: | Intersectin-1, UNKNOWN ATOM OR ION, peptide ligand | | 著者 | Dong, A, Guan, X, Huang, H, Wernimont, A, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2012-12-20 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the Second SH3 Domain of ITSN1 bound with a synthetic peptide
To be Published
|
|
4H9S
 
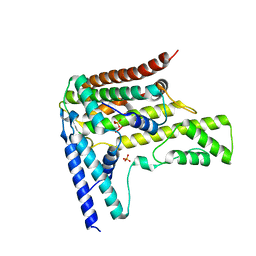 | | Complex structure 6 of DAXX/H3.3(sub7)/H4 | | 分子名称: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | 著者 | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | 登録日 | 2012-09-24 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9P
 
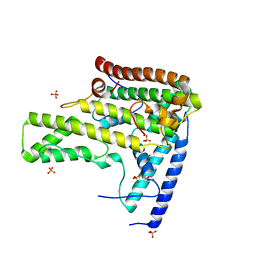 | | Complex structure 3 of DAXX/H3.3(sub5,G90A)/H4 | | 分子名称: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | 著者 | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | 登録日 | 2012-09-24 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9N
 
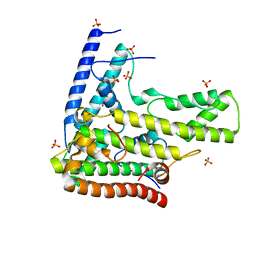 | | Complex structure 1 of DAXX/H3.3(sub5)/H4 | | 分子名称: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | 著者 | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | 登録日 | 2012-09-24 | | 公開日 | 2012-10-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
8Z02
 
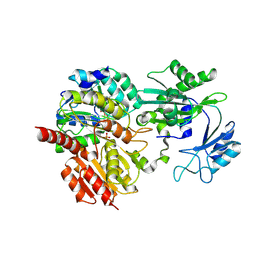 | | CoA bound to human GTP-specific succinyl-CoA synthetase | | 分子名称: | COENZYME A, PHOSPHATE ION, Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | 著者 | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | 登録日 | 2024-04-08 | | 公開日 | 2024-12-11 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
8Z03
 
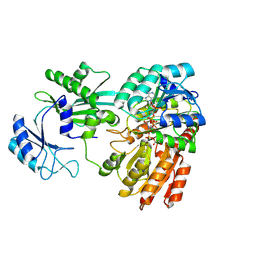 | | Lactate bound to human GTP-specific succinyl-CoA synthetase | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, COENZYME A, MAGNESIUM ION, ... | | 著者 | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | 登録日 | 2024-04-08 | | 公開日 | 2024-12-11 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
7CJM
 
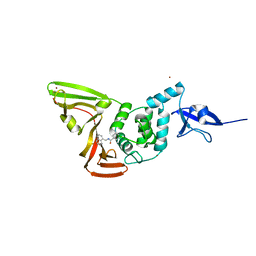 | | SARS CoV-2 PLpro in complex with GRL0617 | | 分子名称: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | 著者 | Fu, Z, Huang, H. | | 登録日 | 2020-07-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
5H2T
 
 | |
6J7B
 
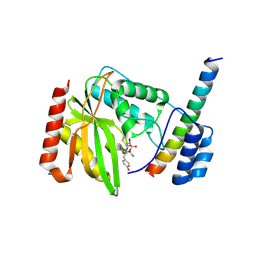 | | Crystal structure of VASH1-SVBP in complex with epoY | | 分子名称: | N-[(3R)-4-ethoxy-3-hydroxy-4-oxobutanoyl]-L-tyrosine, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | 著者 | Wang, N, Bao, H, Huang, H, Wu, B. | | 登録日 | 2019-01-17 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.618 Å) | | 主引用文献 | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
7V1M
 
 | |
6J4P
 
 | |
6J4V
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | 分子名称: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-01-10 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4U
 
 | |
6J4Q
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | 分子名称: | GLYCEROL, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide, Small vasohibin-binding protein, ... | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-01-10 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4S
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | 分子名称: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-01-10 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6KBU
 
 | | Crystal structure of yedK | | 分子名称: | GLYCEROL, SOS response-associated protein | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-06-26 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KIJ
 
 | | Crystal structure of yedK with ssDNA containing an abasic site | | 分子名称: | DNA (5'-D(*GP*AP*TP*TP*CP*GP*TP*CP*G)-3'), GLYCEROL, PENTANE-3,4-DIOL-5-PHOSPHATE, ... | | 著者 | Wang, N, Bao, H, Huang, H. | | 登録日 | 2019-07-18 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KBZ
 
 | |
