7YHG
 
 | |
7YHH
 
 | |
7YHF
 
 | |
7YHI
 
 | |
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | 分子名称: | Nanobody Nb-007, Spike protein S1 | | 著者 | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | 登録日 | 2021-11-20 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
3J4F
 
 | | Structure of HIV-1 capsid protein by cryo-EM | | 分子名称: | capsid protein | | 著者 | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | 登録日 | 2013-07-11 | | 公開日 | 2013-07-24 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J3Y
 
 | |
3J3Q
 
 | |
8HSC
 
 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-19 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS3
 
 | | Gi bound orphan GPR20 in ligand-free state | | 分子名称: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-16 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS2
 
 | | Orphan GPR20 in complex with Fab046 | | 分子名称: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-16 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
4FZ4
 
 | | Crystal structure of HP0197-18kd | | 分子名称: | CHLORIDE ION, NITRATE ION, Uncharacterized protein conserved in bacteria | | 著者 | Yuan, Z, Yan, X. | | 登録日 | 2012-07-06 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Molecular mechanism by which surface antigen HP0197 mediates host cell attachment in the pathogenic bacteria Streptococcus suis
J.Biol.Chem., 288, 2013
|
|
4FZQ
 
 | | Crystal structure of HP0197-G5 | | 分子名称: | Uncharacterized protein conserved in bacteria | | 著者 | Yuan, Z, Yan, X. | | 登録日 | 2012-07-07 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism by which surface antigen HP0197 mediates host cell attachment in the pathogenic bacteria Streptococcus suis
J.Biol.Chem., 288, 2013
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | 著者 | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | 登録日 | 2020-11-22 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | 著者 | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | 登録日 | 2020-11-22 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
4H2A
 
 | |
7X2K
 
 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | 分子名称: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | 分子名称: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | 分子名称: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7Y1F
 
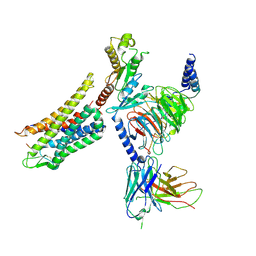 | | Cryo-EM structure of human k-opioid receptor-Gi complex | | 分子名称: | Dynorphin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Chen, B.O, Xu, F.E. | | 登録日 | 2022-06-08 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-06-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of human kappa-opioid receptor-Gi complex bound to an endogenous agonist dynorphin A.
Protein Cell, 14, 2023
|
|
7DPM
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW06 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | 著者 | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | 登録日 | 2020-12-20 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.304 Å) | | 主引用文献 | Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.
Mabs, 13, 2021
|
|
4NAH
 
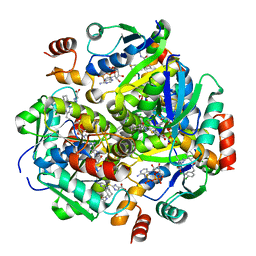 | | Inhibitors of 4-Phosphopanthetheine Adenylyltransferase (PPAT) | | 分子名称: | 2-[(2-{(1S,2S)-2-[(3,4-dichlorobenzyl)carbamoyl]cyclohexyl}-6-ethylpyrimidin-4-yl)sulfanyl]-1H-imidazole-5-carboxylic acid, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phosphopantetheine adenylyltransferase | | 著者 | Lahiri, S.D. | | 登録日 | 2013-10-22 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Discovery of inhibitors of 4'-phosphopantetheine adenylyltransferase (PPAT) to validate PPAT as a target for antibacterial therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
7VAA
 
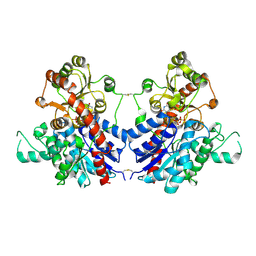 | |
7VA8
 
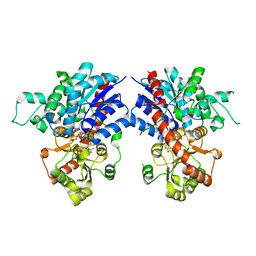 | | Crystal structure of MiCGT | | 分子名称: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Zhong, L, Zhang, Z.M. | | 登録日 | 2021-08-27 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85003233 Å) | | 主引用文献 | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
