5I8E
 
 | | Crystal Structure of Broadly Neutralizing HIV-1 Fusion Peptide-Targeting Antibody VRC34.01 Fab | | 分子名称: | VRC34.01 Fab heavy chain, VRC34.01 Fab light chain, ZINC ION | | 著者 | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | 登録日 | 2016-02-18 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.655 Å) | | 主引用文献 | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
5I8C
 
 | | Crystal Structure of HIV-1 Clade A BG505 Fusion Peptide (residue 512-520) in Complex with Broadly Neutralizing Antibody VRC34.01 Fab | | 分子名称: | HIV-1 Clade A BG505 Fusion Peptide (residue 512-520), VRC34.01 Fab heavy chain, VRC34.01 Fab light chain | | 著者 | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | 登録日 | 2016-02-18 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
6ISL
 
 | | SnoaL-like cyclase XimE with its product xiamenmycin B | | 分子名称: | (2R,3S)-2-methyl-2-(4-methylpent-3-enyl)-3-oxidanyl-3,4-dihydrochromene-6-carboxylic acid, XimE, SnoaL-like domain protein | | 著者 | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | 登録日 | 2018-11-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
6ISK
 
 | | SnoaL-like cyclase XimE | | 分子名称: | 1,2-ETHANEDIOL, MALONIC ACID, XimE, ... | | 著者 | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | 登録日 | 2018-11-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
4RWY
 
 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC131 in complex with HIV-1 clade B YU2 gp120 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Acharya, P, Luongo, T.S, Kwong, P.D. | | 登録日 | 2014-12-08 | | 公開日 | 2015-07-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.128 Å) | | 主引用文献 | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
2GCD
 
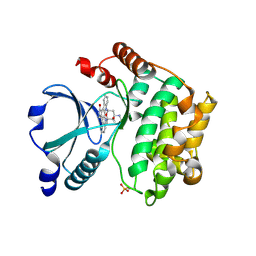 | | TAO2 kinase domain-staurosporine structure | | 分子名称: | STAUROSPORINE, Serine/threonine-protein kinase TAO2 | | 著者 | Zhou, T, Sun, L, Gao, Y, Earnest, S, Cobb, M.H, Goldsmith, E.J. | | 登録日 | 2006-03-14 | | 公開日 | 2006-09-05 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of the MAP3K TAO2 kinase domain bound by an inhibitor staurosporine.
Acta Biochim.Biophys.Sinica, 38, 2006
|
|
7KNI
 
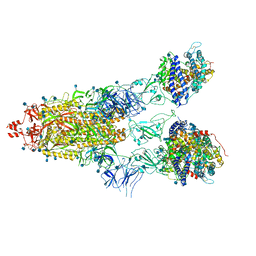 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNE
 
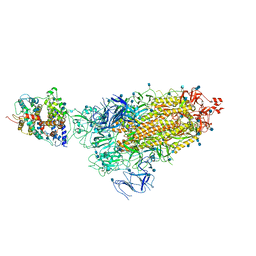 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
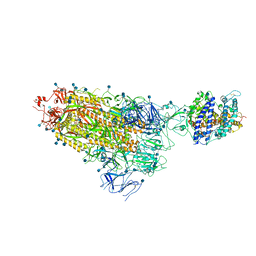 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMB
 
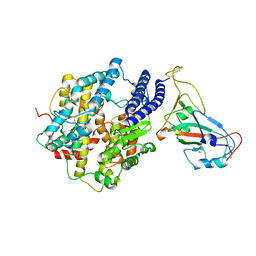 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-02 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
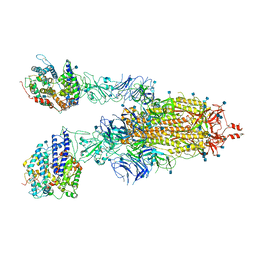 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMS
 
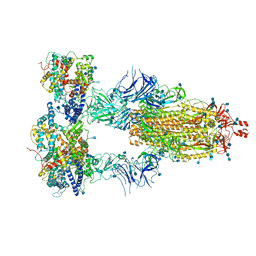 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
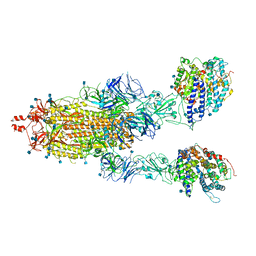 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7LPN
 
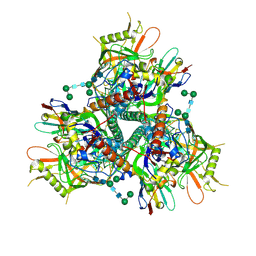 | |
8TOP
 
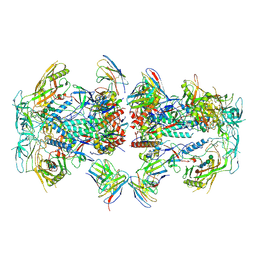 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | 著者 | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D. | | 登録日 | 2023-08-03 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide
To Be Published
|
|
8F6X
 
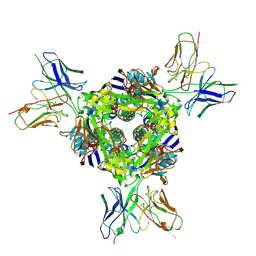 | | cryo-EM structure of a structurally designed Human metapneumovirus F protein in complex with antibody MPE8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MPE8 Single chain variable fragment, Structurally designed HMPV F protein HMPV_v3B_D12_DS454,Fibritin | | 著者 | Zhou, T, Kwong, P.D, Morano, N.C, Ou, L. | | 登録日 | 2022-11-17 | | 公開日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | cryo-EM structure of a structurally designed Human metapneumovirus F protein in complex with antibody MPE8
To Be Published
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | 分子名称: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rapp, M, Shapiro, L, Ho, D.D. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
6E5P
 
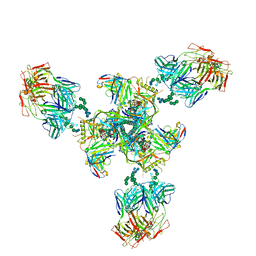 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | 分子名称: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | 著者 | Acharya, P, Kwong, P.D. | | 登録日 | 2018-07-21 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
7MY2
 
 | |
7MY3
 
 | |
4MMS
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 5.5 | | 分子名称: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, SULFATE ION | | 著者 | Mclellan, J.S, Joyce, M.G, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | 登録日 | 2013-09-09 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMR
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 9.5 | | 分子名称: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | 著者 | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | 登録日 | 2013-09-09 | | 公開日 | 2013-11-20 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMV
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1-TriC at pH 9.5 | | 分子名称: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | 著者 | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | 登録日 | 2013-09-09 | | 公開日 | 2013-11-20 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
