4HDP
 
 | | Crystal Structure of HIV-1 protease mutants I50V complexed with inhibitor GRL-0519 | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | 著者 | Shen, C.H, Zhang, H, Weber, I.T. | | 登録日 | 2012-10-02 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HE9
 
 | | Crystal Structure of HIV-1 protease mutants I54M complexed with inhibitor GRL-0519 | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 protease, ... | | 著者 | Shen, C.H, Zhang, H, Weber, I.T. | | 登録日 | 2012-10-03 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
3PWM
 
 | | HIV-1 Protease Mutant L76V with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3NU6
 
 | |
3NUJ
 
 | |
3NUO
 
 | |
3NU9
 
 | |
4L13
 
 | |
4J5J
 
 | |
4L1I
 
 | |
4L12
 
 | |
6B4N
 
 | | a hydroxymethyl functionality at the 4-position of the 2-phenyloxazole moiety of HIV-1 protease inhibitors involving the P2' ligands | | 分子名称: | CHLORIDE ION, Protease, SODIUM ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-09-27 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2' Ligands of Darunavir.
ChemMedChem, 12, 2017
|
|
7EKO
 
 | | CrClpP-S1 | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKQ
 
 | | CrClpP-S2c | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
2G69
 
 | |
6BKA
 
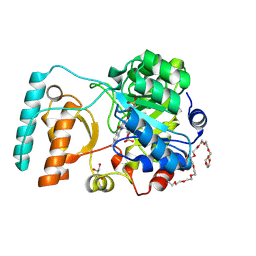 | | Crystal Structure of Nitronate Monooxygenase from Cyberlindnera saturnus | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Agniswamy, J, Fang, Y.-F, Weber, I.T. | | 登録日 | 2017-11-08 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of yeast nitronate monooxygenase from Cyberlindnera saturnus.
Proteins, 86, 2018
|
|
7WVQ
 
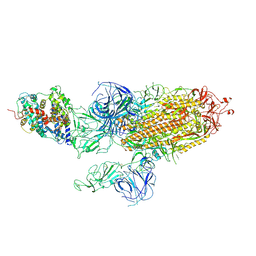 | |
7WVP
 
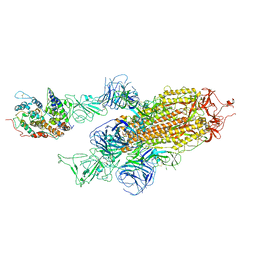 | |
7WK5
 
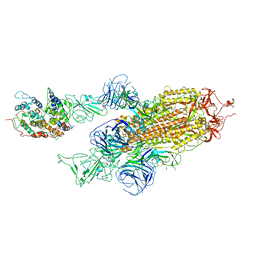 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Han, W.Y, Wang, Y.F. | | 登録日 | 2022-01-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-03-09 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK6
 
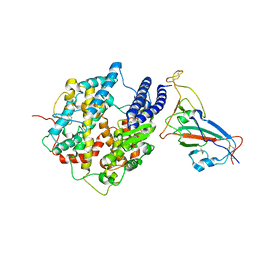 | |
7WK4
 
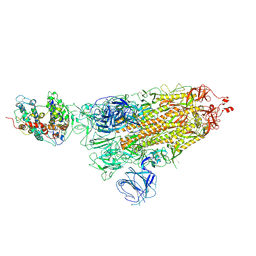 | |
3D1X
 
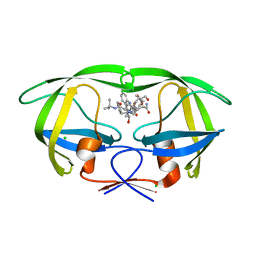 | | Crystal structure of HIV-1 mutant I54M and inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, F, Weber, I.T. | | 登録日 | 2008-05-06 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3CYW
 
 | | Effect of Flap Mutations on Structure of HIV-1 Protease and Inhibition by Saquinavir and Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, F, Weber, I.T. | | 登録日 | 2008-04-27 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D1Y
 
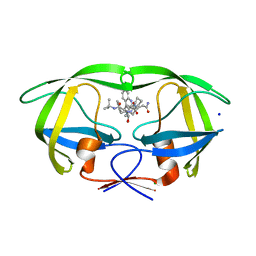 | | Crystal structure of HIV-1 mutant I54V and inhibitor SAQUINA | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 Protease, ... | | 著者 | Liu, F, Weber, I.T. | | 登録日 | 2008-05-06 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
