8Q93
 
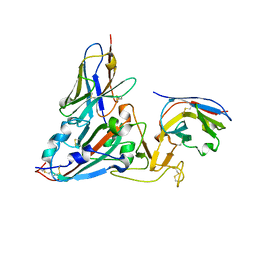 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | 分子名称: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | 著者 | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | 登録日 | 2023-08-19 | | 公開日 | 2023-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q95
 
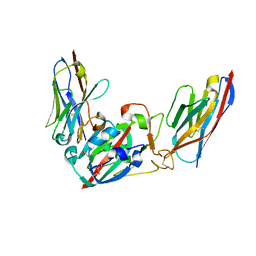 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | 分子名称: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | 著者 | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | 登録日 | 2023-08-19 | | 公開日 | 2023-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q94
 
 | |
7OLZ
 
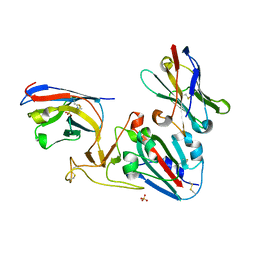 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | 分子名称: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | 著者 | Aksu, M, Guttler, T, Gorlich, D. | | 登録日 | 2021-05-20 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7ON5
 
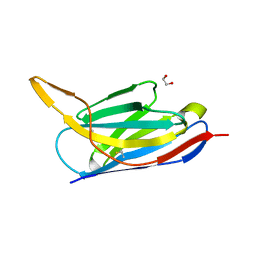 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | 著者 | Aksu, M, Guttler, T, Gorlich, D. | | 登録日 | 2021-05-25 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7P98
 
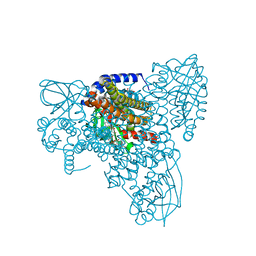 | | Cyclohex-1-ene-1-carboxyl-CoA dehydrogenase in a substrate-free state | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Short-chain acyl-CoA dehydrogenase | | 著者 | Ermler, U, Weidenweber, S, Boll, M. | | 登録日 | 2021-07-26 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
7P9X
 
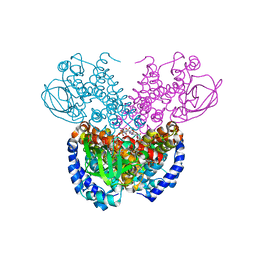 | | Structure of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase complexed with cyclohex-1-ene-1-carboxyl-CoA | | 分子名称: | 1-monoenoyl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Ermler, U, Weidenweber, S, Boll, M. | | 登録日 | 2021-07-28 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
7P9A
 
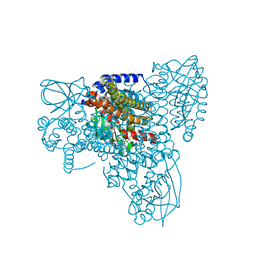 | | Structure of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase complexed with cyclohex-1,5-diene-1-carboxyl-CoA | | 分子名称: | 1,5 Dienoyl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Ermler, U, Weidenweber, S, Boll, M. | | 登録日 | 2021-07-26 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
4Z3L
 
 | |
6RV2
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | 著者 | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-05-30 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6SPX
 
 | | Structure of protein kinase CK2 catalytic subunit in complex with the CK2beta-competitive bisubstrate inhibitor ARC1502 | | 分子名称: | 8-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]octanoic acid, ARC1502, Casein kinase II subunit alpha | | 著者 | Niefind, K, Schnitzler, A. | | 登録日 | 2019-09-03 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
6RV4
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | 著者 | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-05-30 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6RV3
 
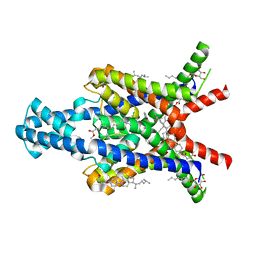 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | 著者 | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-05-30 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6SPW
 
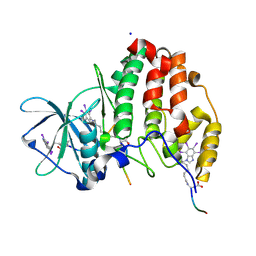 | | Structure of protein kinase CK2 catalytic subunit with the CK2beta-competitive bisubstrate inhibitor ARC3140 | | 分子名称: | 8-[4,5,6,7-tetrakis(iodanyl)benzimidazol-1-yl]octanoic acid, ARC3140, Casein kinase II subunit alpha, ... | | 著者 | Niefind, K, Schnitzler, A. | | 登録日 | 2019-09-03 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
7XAD
 
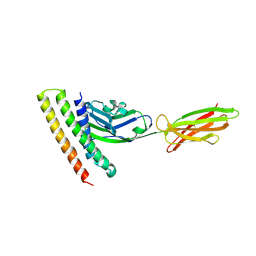 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | 分子名称: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | 著者 | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | 登録日 | 2022-03-17 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
1AUU
 
 | |
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-08 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
2I2S
 
 | |
8F73
 
 | |
3FH5
 
 | |
3FH8
 
 | |
2DWR
 
 | |
1S6P
 
 | |
