1YDE
 
 | | Crystal Structure of Human Retinal Short-Chain Dehydrogenase/Reductase 3 | | 分子名称: | Retinal dehydrogenase/reductase 3 | | 著者 | Lukacik, P, Bunkozci, G, Kavanagh, K, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2004-12-23 | | 公開日 | 2005-01-18 | | 最終更新日 | 2012-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity.
Biochem.J., 402, 2007
|
|
2XHS
 
 | |
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 2004-05-30 | | 公開日 | 2005-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W01
 
 | |
1W02
 
 | |
8E1O
 
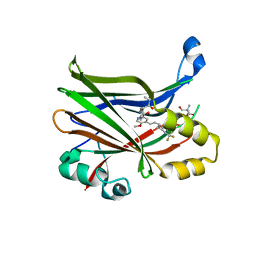 | | Crystal structure of hTEAD2 bound to a methoxypyridine lipid pocket binder | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methoxy-N-({3-[2-(methylamino)-2-oxoethyl]phenyl}methyl)-4-{(E)-2-[trans-4-(trifluoromethyl)cyclohexyl]ethenyl}pyridine-2-carboxamide, Transcriptional enhancer factor TEF-4 | | 著者 | Noland, C.L, Dey, A, Zbieg, J, Crawford, J. | | 登録日 | 2022-08-10 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Targeting the Hippo pathway in cancers via ubiquitination dependent TEAD degradation
Biorxiv, 2024
|
|
1NE3
 
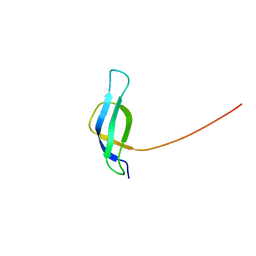 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | 分子名称: | 30S ribosomal protein S28E | | 著者 | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-12-10 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
4MLE
 
 | |
4MLH
 
 | |
6DTW
 
 | | HIV-1 Reverse Transcriptase Y181C Mutant in complex with JLJ 578 | | 分子名称: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | 登録日 | 2018-06-18 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.742 Å) | | 主引用文献 | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7TYQ
 
 | | TEAD2 bound to Compound 1 | | 分子名称: | Transcriptional enhancer factor TEF-4, ethyl (8S)-7-oxo-5-[4-(trifluoromethyl)phenyl]-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carboxylate | | 著者 | Noland, C.L, Fong, R. | | 登録日 | 2022-02-14 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYU
 
 | | TEAD2 bound to Compound 2 | | 分子名称: | (3R)-1-[(8S)-5-(4-cyclohexylphenyl)-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonyl]pyrrolidine-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | 著者 | Noland, C.L, Fong, R. | | 登録日 | 2022-02-14 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYP
 
 | | TEAD2 bound to GNE-7883 | | 分子名称: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | 著者 | Noland, C.L, Fong, R. | | 登録日 | 2022-02-14 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
6DTX
 
 | | Wildtype HIV-1 Reverse Transcriptase in complex with JLJ 578 | | 分子名称: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, GLYCEROL, Reverse transcriptase/ribonuclease H, ... | | 著者 | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | 登録日 | 2018-06-18 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.327 Å) | | 主引用文献 | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5EJJ
 
 | | Crystal structure of UfSP from C.elegans | | 分子名称: | Ufm1-specific protease | | 著者 | Kim, K, Ha, B, Kim, E.E. | | 登録日 | 2015-11-02 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
6AK2
 
 | | Crystal structure of the syntenin PDZ1 domain in complex with the peptide inhibitor KSL-128018 | | 分子名称: | Syntenin-1, peptide inhibitor KSL-128018 | | 著者 | Jin, Z.Y, Park, J.H, Yun, J.H, Haugaard-Kedstrom, L.M, Lee, W.T. | | 登録日 | 2018-08-29 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.868 Å) | | 主引用文献 | A High-Affinity Peptide Ligand Targeting Syntenin Inhibits Glioblastoma.
J.Med.Chem., 64, 2021
|
|
5DIA
 
 | | PIM1 in complex with Cpd36 ((1S,3S)-N1-(6-(5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridin-3-yl)pyridin-2-yl)cyclohexane-1,3-diamine) | | 分子名称: | (1S,3S)-N-{6-[5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridin-3-yl]pyridin-2-yl}cyclohexane-1,3-diamine, Serine/threonine-protein kinase pim-1 | | 著者 | Murray, J.M, Wallweber, H. | | 登録日 | 2015-08-31 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.964 Å) | | 主引用文献 | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
