7UG8
 
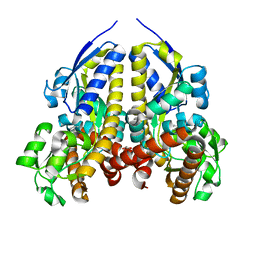 | | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium | | 分子名称: | 1,2-ETHANEDIOL, 2-oxopentanoic acid, CALCIUM ION, ... | | 著者 | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | 登録日 | 2022-03-24 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium
To Be Published
|
|
6YM0
 
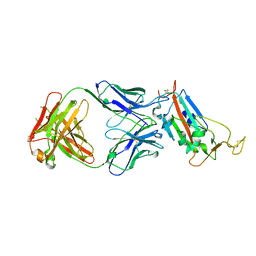 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | 分子名称: | Spike glycoprotein, heavy chain, light chain | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.36 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | 分子名称: | IgG H chain, IgG L chain, Spike glycoprotein | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-15 | | 公開日 | 2020-04-29 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
5EYT
 
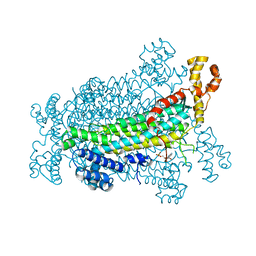 | | Crystal Structure of Adenylosuccinate Lyase from Schistosoma mansoni in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase | | 著者 | Romanello, L, Torini, J.R, Bird, L, Nettleship, J, Owens, R, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | 登録日 | 2015-11-25 | | 公開日 | 2016-11-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3649 Å) | | 主引用文献 | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
6YLA
 
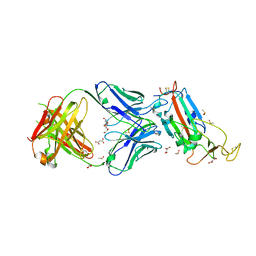 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-06 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
7OAQ
 
 | |
7OAO
 
 | | Nanobody C5 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAP
 
 | | Nanobody H3 AND C1 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAN
 
 | | Nanobody C5 bound to Spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Naismith, J.H, Weckener, M. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAY
 
 | | Nanobody F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-20 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | |
7Z1A
 
 | | Nanobody H11 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 Nanobody, H11 Nanobody, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1D
 
 | | Nanobody H11-H6 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H6 nanobody, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1C
 
 | | Nanobody H11-B5 and H11-F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Nanobody B5, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1B
 
 | | Nanobody H11-A10 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1E
 
 | | Nanobody H11-H4 Q98R H100E bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H4 Q98R H100E, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z85
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9Q
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z7X
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z86
 
 | |
7Z6V
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | 著者 | Weckener, M, Naismith, J.H, Vogirala, V.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9R
 
 | |
6SIR
 
 | |
6SNK
 
 | | Crystal structure of the Collagen VI alpha3 N2 domain | | 分子名称: | Collagen alpha-3(VI) chain | | 著者 | Gebauer, J.M, Degefa, H.S, Paulsson, M, Wagener, R, Baumann, U. | | 登録日 | 2019-08-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a collagen VI alpha 3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations.
J.Biol.Chem., 295, 2020
|
|
6ZBP
 
 | | H11-H4 complex with SARS-CoV-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H11-H4, SULFATE ION, ... | | 著者 | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, LeBas, A. | | 登録日 | 2020-06-08 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
