4QJE
 
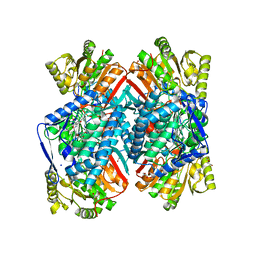 | | 1.85 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-free sulfinic acid form of Cys289 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-06-03 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2HKY
 
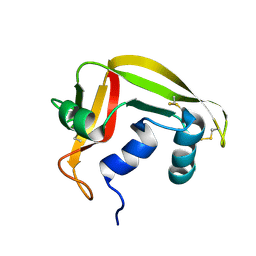 | |
2K9N
 
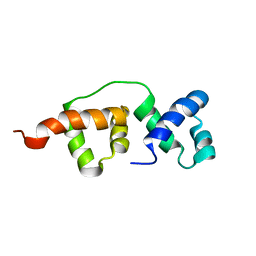 | | Solution NMR structure of the R2R3 DNA binding domain of Myb1 protein from protozoan parasite Trichomonas vaginalis | | 分子名称: | MYB24 | | 著者 | Lou, Y, Wei, S, Rajasekaran, M, Chou, C, Hsu, H, Tai, J, Chen, C. | | 登録日 | 2008-10-19 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 37, 2009
|
|
3ZQC
 
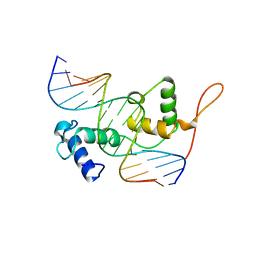 | | Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal beta-hairpin conformation | | 分子名称: | MRE-1, MYB3 | | 著者 | Wei, S.-Y, Lou, Y.-C, Tsai, J.-Y, Hsu, H.-M, Tai, J.-H, Hsiao, C.-D, Chen, C. | | 登録日 | 2011-06-09 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the Trichomonas Vaginalis Myb3 DNA-Binding Domain Bound to a Promoter Sequence Reveals a Unique C-Terminal Beta-Hairpin Conformation.
Nucleic Acids Res., 40, 2012
|
|
4HND
 
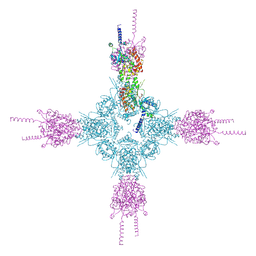 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | 著者 | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | 登録日 | 2012-10-19 | | 公開日 | 2014-04-09 | | 最終更新日 | 2016-12-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
5GWY
 
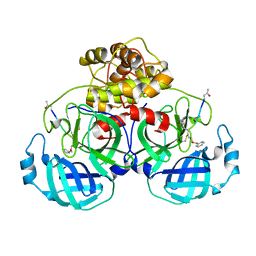 | | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design | | 分子名称: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, main protease | | 著者 | Wang, F, Chen, C, Tan, W, Yang, K, Yang, H. | | 登録日 | 2016-09-14 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.852 Å) | | 主引用文献 | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design.
Sci Rep, 6, 2016
|
|
5GJB
 
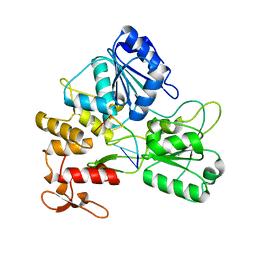 | | Zika virus NS3 helicase in complex with ssRNA | | 分子名称: | NS3 helicase, RNA (5'-R(*AP*GP*AP*UP*CP*AP*A)-3') | | 著者 | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | 登録日 | 2016-06-28 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
5GJC
 
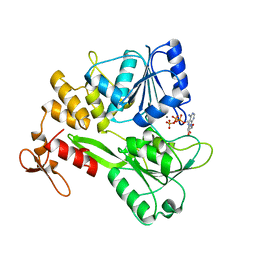 | | Zika virus NS3 helicase in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, NS3 helicase | | 著者 | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | 登録日 | 2016-06-28 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.204 Å) | | 主引用文献 | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
5H2S
 
 | |
4HNE
 
 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | 著者 | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | 登録日 | 2012-10-19 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
2M87
 
 | |
2JXU
 
 | |
2KDZ
 
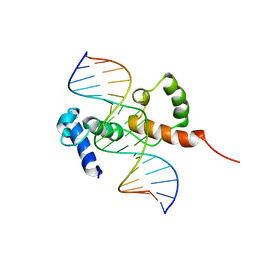 | | Structure of the R2R3 DNA binding domain of MYB1 protein from protozoan parasite trichomonas vaginalis in complex with MRE-1/MRE-2R DNA | | 分子名称: | 5'-D(*AP*AP*GP*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*TP*A)-3', 5'-D(*TP*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*TP*CP*TP*T)-3', MYB24 | | 著者 | Lou, Y.C, Wei, S.Y, Rajasekaran, M, Chou, C.C, Hsu, H.M, Tai, J.H, Chen, C. | | 登録日 | 2009-01-21 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 2009
|
|
2H6L
 
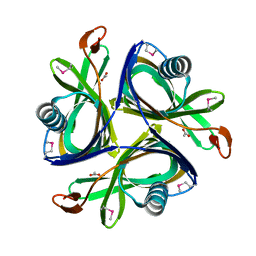 | | X-Ray Crystal Structure of the Metal-containing Protein AF0104 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium Target GR103. | | 分子名称: | ACETIC ACID, Hypothetical protein, ZINC ION | | 著者 | Kuzin, A.P, Abashidze, M, Fang, Y, Chen, C, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-05-31 | | 公開日 | 2006-07-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Three dimensional structure of the hypothetical protein
AF0104 at the 2.0 A resolution.
To be Published
|
|
2KVH
 
 | |
2KXV
 
 | |
2KVF
 
 | |
2KXT
 
 | |
2KVG
 
 | |
2KV3
 
 | |
4Q92
 
 | | 1.90 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-modified Cys289 | | 分子名称: | Betaine aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-04-28 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3GCX
 
 | | PCSK9:EGFA (pH 7.4) | | 分子名称: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | 著者 | McNutt, M.C, Kwon, H.J, Chen, C, Chen, J.R, Horton, J.D, Lagace, T.A. | | 登録日 | 2009-02-22 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Antagonism of Secreted PCSK9 Increases Low Density Lipoprotein Receptor Expression in HepG2 Cells.
J.Biol.Chem., 284, 2009
|
|
5JMT
 
 | | Crystal structure of Zika virus NS3 helicase | | 分子名称: | NS3 helicase | | 著者 | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | 登録日 | 2016-04-29 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | 分子名称: | ZIKV NS1 | | 著者 | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | 登録日 | 2017-03-03 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.817 Å) | | 主引用文献 | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
5I9Q
 
 | | Crystal structure of 3BNC55 Fab in complex with 426c.TM4deltaV1-3 gp120 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC55 Fab heavy chain, ... | | 著者 | Scharf, L, Chen, C, Bjorkman, P.J. | | 登録日 | 2016-02-20 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
