5L0A
 
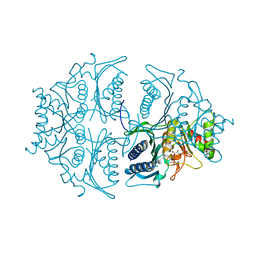 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-1,6-bisphosphate | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | 著者 | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | 登録日 | 2016-07-27 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Structural studies of human muscle FBPase
To Be Published
|
|
6UR7
 
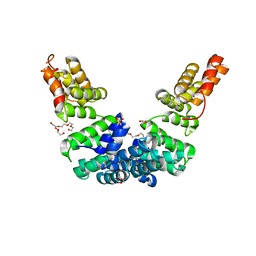 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | 著者 | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2019-10-22 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.709 Å) | | 主引用文献 | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
2M5R
 
 | |
6USV
 
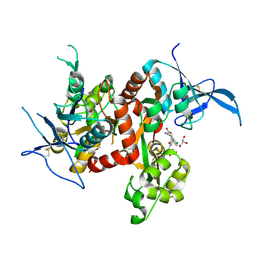 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | 分子名称: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | 著者 | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6V72
 
 | | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase II, CALCIUM ION, ... | | 著者 | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-06 | | 公開日 | 2019-12-25 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis
To Be Published
|
|
2M20
 
 | | EGFR transmembrane - juxtamembrane (TM-JM) segment in bicelles: MD guided NMR refined structure. | | 分子名称: | Epidermal growth factor receptor | | 著者 | Endres, N.F, Das, R, Smith, A, Arkhipov, A, Kovacs, E, Huang, Y, Pelton, J.G, Shan, Y, Shaw, D.E, Wemmer, D.E, Groves, J.T, Kuriyan, J. | | 登録日 | 2012-12-11 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor.
Cell(Cambridge,Mass.), 152, 2013
|
|
7S83
 
 | | Crystal structure of SARS CoV-2 Spike Receptor Binding Domain in complex with shark neutralizing VNARs ShAb01 and ShAb02 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ShAb01 VNAR, ... | | 著者 | Chen, W.-H, Hajduczki, A, Dooley, H.M, Joyce, M.G. | | 登録日 | 2021-09-17 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity.
Nat Commun, 14, 2023
|
|
6VCG
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | 分子名称: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | 著者 | Daruwalla, A, Shi, W, Kiser, P.D. | | 登録日 | 2019-12-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | 分子名称: | Muramoyltetrapeptide carboxypeptidase | | 著者 | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2019-10-30 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.348 Å) | | 主引用文献 | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
2MJ9
 
 | | Designed Exendin-4 analogues | | 分子名称: | Exendin-4 | | 著者 | Rovo, P, Farkas, V, Straner, P, Szabo, M, Jermendy, A, Hegyi, O, Toth, G.K, Perczel, A. | | 登録日 | 2013-12-30 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational design of alpha-helix-stabilized exendin-4 analogues.
Biochemistry, 53, 2014
|
|
6UPG
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with cYF-4-OMe | | 分子名称: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(4-methoxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nguyen, R.C.D, Yang, Y, Liu, A. | | 登録日 | 2019-10-17 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Substrate-Assisted Hydroxylation and O-Demethylation in the Peroxidase-like Cytochrome P450 Enzyme CYP121
Acs Catalysis, 10, 2020
|
|
6UPT
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706 | | 分子名称: | 2-((2-chlorobenzyl)thio)-4,5-dihydro-1H-imidazole, TP53-binding protein 1, UNKNOWN ATOM OR ION | | 著者 | The, J, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-18 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706
to be published
|
|
7SA5
 
 | | Two-state solution NMR structure of Apo Pin1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Born, A, Vogeli, B. | | 登録日 | 2021-09-22 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Reconstruction of Coupled Intra- and Interdomain Protein Motion from Nuclear and Electron Magnetic Resonance.
J.Am.Chem.Soc., 143, 2021
|
|
7SJL
 
 | |
7T8Q
 
 | |
7T8P
 
 | |
6V11
 
 | | Lon Protease from Yersinia pestis | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | 著者 | Shin, M, Puchades, C, Asmita, A, Puri, N, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | 登録日 | 2019-11-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
1U5A
 
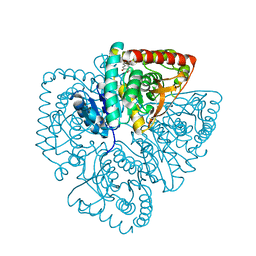 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,5-dihydroxy-2-naphthoic acid | | 分子名称: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase | | 著者 | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | 登録日 | 2004-07-27 | | 公開日 | 2005-06-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
6ZUP
 
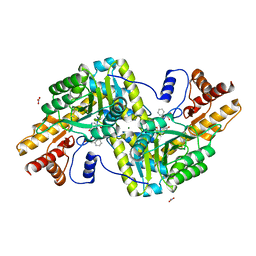 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-(-)-3-phenyllactic acid | | 分子名称: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, Aminotransferase, MAGNESIUM ION, ... | | 著者 | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | 登録日 | 2020-07-23 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUR
 
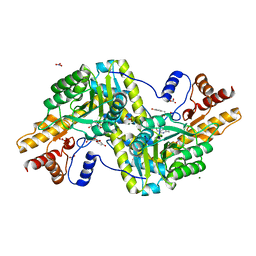 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-p-hydroxyphenyllactic acid | | 分子名称: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, Aminotransferase, MAGNESIUM ION, ... | | 著者 | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | 登録日 | 2020-07-23 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
1UH1
 
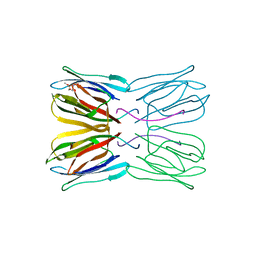 | | Crystal structure of jacalin- GalNAc-beta(1-3)-Gal-alpha-O-Me complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-methyl alpha-D-galactopyranoside, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | 著者 | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | 登録日 | 2003-06-23 | | 公開日 | 2003-09-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
7SQE
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with Jun9-84-3 inhibitor | | 分子名称: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-11-05 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with Jun9-84-3 inhibitor
To be Published
|
|
7SZ7
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha. "tips-juxtaposed" conformation | | 分子名称: | Epidermal growth factor receptor, Transforming growth factor alpha | | 著者 | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | 登録日 | 2021-11-25 | | 公開日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ1
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | 分子名称: | Epidermal growth factor, Epidermal growth factor receptor | | 著者 | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | 登録日 | 2021-11-25 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYD
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | 分子名称: | Epidermal growth factor, Epidermal growth factor receptor | | 著者 | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | 登録日 | 2021-11-24 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
