8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.36 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6UEG
 
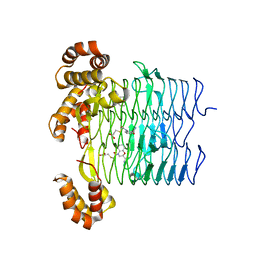 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | 分子名称: | 3-({2-[(2R)-2-carbamoyl-2,3-dihydro-4H-1,4-benzoxazin-4-yl]-2-oxoethyl}sulfanyl)propanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CALCIUM ION | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
6UEC
 
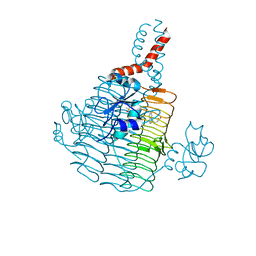 | | Pseudomonas aeruginosa LpxD Complex Structure with Ligand | | 分子名称: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UEE
 
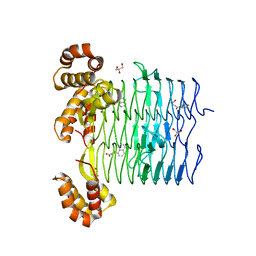 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | 分子名称: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, GLYCEROL | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
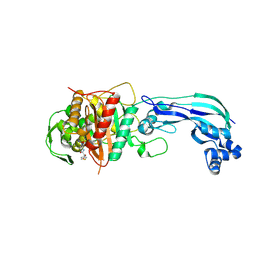 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | 分子名称: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UNB
 
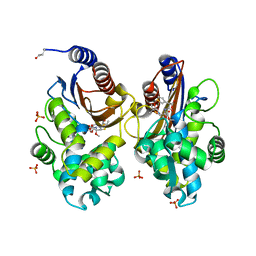 | | Crystal structure of CTX-M-14 in complex with temocillin | | 分子名称: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-11 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UED
 
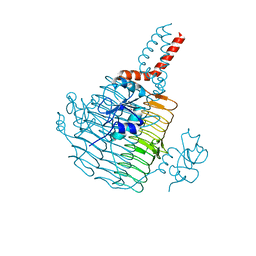 | | Apo Pseudomonas aeruginosa LpxD Structure | | 分子名称: | GLYCEROL, MAGNESIUM ION, UDP-3-O-acylglucosamine N-acyltransferase | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
2FHS
 
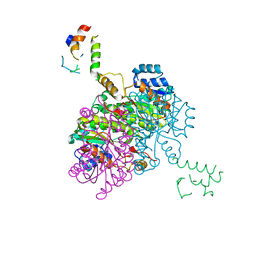 | | Structure of Acyl Carrier Protein Bound to FabI, the Enoyl Reductase from Escherichia Coli | | 分子名称: | Acyl carrier protein, enoyl-[acyl-carrier-protein] reductase, NADH-dependent | | 著者 | Kolappan, S, Novichenok, P, Rafi, S, Simmerling, C, Tonge, P.J, Kisker, C. | | 登録日 | 2005-12-27 | | 公開日 | 2006-10-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Acyl Carrier Protein Bound to FabI, the FASII Enoyl Reductase from Escherichia coli.
J.Biol.Chem., 281, 2006
|
|
5ZUG
 
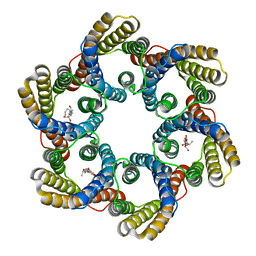 | | Structure of the bacterial acetate channel SatP | | 分子名称: | Succinate-acetate/proton symporter SatP, nonyl beta-D-glucopyranoside | | 著者 | Sun, P.C, Li, J.L, Xiao, Q.J, Guan, Z.Y, Deng, D. | | 登録日 | 2018-05-07 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel.
J. Biol. Chem., 293, 2018
|
|
2FDW
 
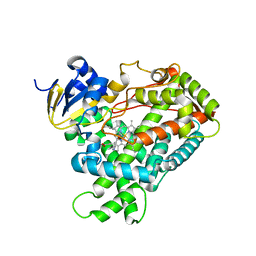 | | Crystal Structure Of Human Microsomal P450 2A6 with the inhibitor (5-(Pyridin-3-yl)furan-2-yl)methanamine bound | | 分子名称: | (5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2L42
 
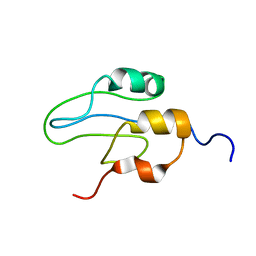 | |
2FDU
 
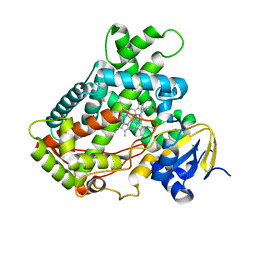 | | Microsomal P450 2A6 with the inhibitor N,N-Dimethyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | 分子名称: | Cytochrome P450 2A6, N,N-DIMETHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2L7E
 
 | |
2FDV
 
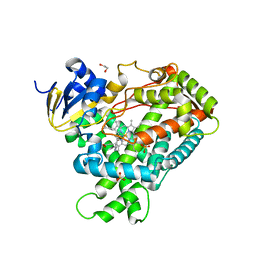 | | Microsomal P450 2A6 with the inhibitor N-Methyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome P450 2A6, N-METHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, ... | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FDY
 
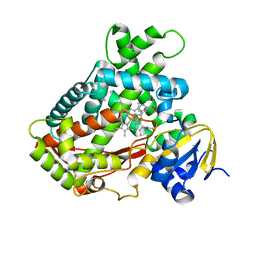 | | Microsomal P450 2A6 with the inhibitor Adrithiol bound | | 分子名称: | 4,4'-DIPYRIDYL DISULFIDE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
6KWC
 
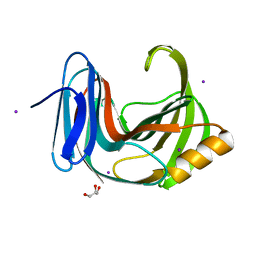 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II | | 分子名称: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | 著者 | Li, C, Wan, Q. | | 登録日 | 2019-09-06 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
8X2J
 
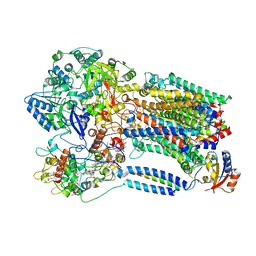 | |
1AKN
 
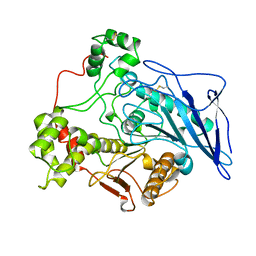 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | 著者 | Wang, X, Zhang, X. | | 登録日 | 1997-05-23 | | 公開日 | 1998-05-27 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
7EKQ
 
 | | CrClpP-S2c | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
