7XWL
 
 | | structure of patulin-detoxifying enzyme Y155F/V187F with NADPH | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWN
 
 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
5Z93
 
 | |
6C26
 
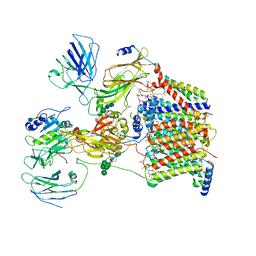 | | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex | | 分子名称: | (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, ... | | 著者 | Bai, L, Li, H. | | 登録日 | 2018-01-06 | | 公開日 | 2018-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The atomic structure of a eukaryotic oligosaccharyltransferase complex.
Nature, 555, 2018
|
|
6C31
 
 | | Crystal structure of TetR family protein Rv0078 in complex with DNA | | 分子名称: | DNA (5'-D(*GP*TP*TP*AP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*CP*TP*TP*GP*TP*AP*AP*A)-3'), DNA (5'-D(P*AP*CP*AP*AP*GP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*TP*AP*AP*C)-3'), TetR family transcriptional regulator | | 著者 | Hsu, H.C, Li, H. | | 登録日 | 2018-01-09 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Cytokinin Signaling in Mycobacterium tuberculosis.
MBio, 9, 2018
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-19 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-18 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-19 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
7VH5
 
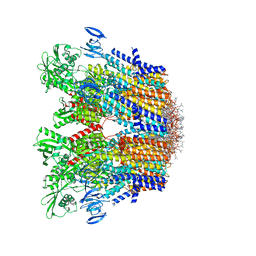 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | 著者 | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | 著者 | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-24 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
6KDR
 
 | |
8GQD
 
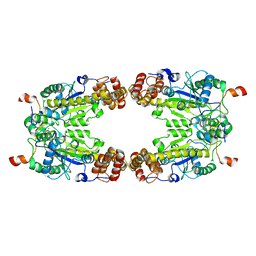 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | 分子名称: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | 著者 | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | 登録日 | 2022-08-30 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Complex Structure and Activation Mechanism of Arginine Kinase McsB by McsA
To Be Published
|
|
6KDQ
 
 | |
6KDS
 
 | |
8IOD
 
 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | 著者 | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | 登録日 | 2023-03-10 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8INR
 
 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | 著者 | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | 登録日 | 2023-03-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8IOC
 
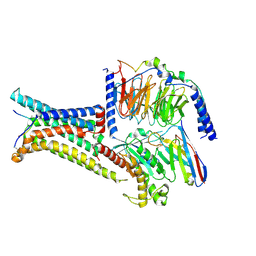 | | Cryo-EM structure of the gamma-MSH-bound human melanocortin receptor 3 (MC3R)-Gs complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | 著者 | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S. | | 登録日 | 2023-03-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
6LK9
 
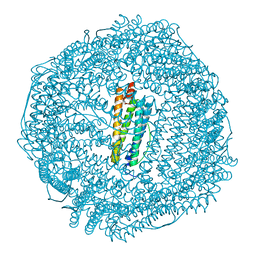 | | Coho salmon ferritin | | 分子名称: | Coho salmon ferritin, FE (III) ION | | 著者 | Wang, Z, Zang, J, Li, H, Tan, X, Du, M. | | 登録日 | 2019-12-18 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Coho salmon ferritin
To Be Published
|
|
7X2U
 
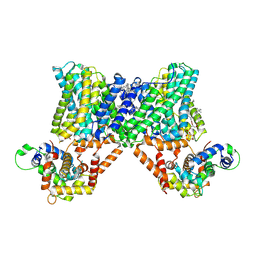 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | 著者 | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | 登録日 | 2022-02-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
7E9M
 
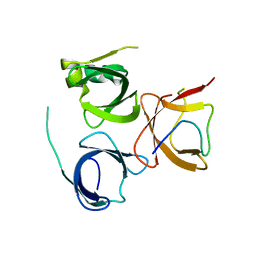 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep3 | | 分子名称: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | 著者 | Zhao, F, Li, H. | | 登録日 | 2021-03-04 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
7EA1
 
 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep2 | | 分子名称: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | 著者 | Zhao, F, Li, H. | | 登録日 | 2021-03-05 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
7BQZ
 
 | |
7BU9
 
 | |
1LTE
 
 | | STRUCTURE OF A LEGUME LECTIN WITH AN ORDERED N-LINKED CARBOHYDRATE IN COMPLEX WITH LACTOSE | | 分子名称: | CALCIUM ION, CORAL TREE LECTIN, MANGANESE (II) ION, ... | | 著者 | Shaanan, B, Lis, H, Sharon, N. | | 登録日 | 1991-06-25 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a legume lectin with an ordered N-linked carbohydrate in complex with lactose.
Science, 254, 1991
|
|
