6YVY
 
 | |
6YXV
 
 | |
6YVS
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 5-{4-[(Pyridin-3-ylmethyl)-amino]-5-trifluoromethyl-pyrimidin-2-ylamino}-1,3-dihydro-indol-2-one | | 分子名称: | 5-[[4-(pyridin-3-ylmethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-1,3-dihydroindol-2-one, Focal adhesion kinase 1, SULFATE ION | | 著者 | Musil, D, Heinrich, T, Amaral, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
6E1Y
 
 | |
6E23
 
 | |
6E22
 
 | |
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | 分子名称: | Surface layer protein | | 著者 | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | 登録日 | 2002-02-12 | | 公開日 | 2002-11-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
6E1Z
 
 | |
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | 分子名称: | Chloroplast pentatricopeptide repeat protein 10 | | 著者 | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | 登録日 | 2013-08-08 | | 公開日 | 2013-10-30 | | 最終更新日 | 2013-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
7L8O
 
 | | OXA-48 bound by Compound 4.3 | | 分子名称: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
8IYD
 
 | | Tail cap of phage lambda tail | | 分子名称: | Tail tube protein, Tail tube terminator protein | | 著者 | Wang, J.W, Wang, C. | | 登録日 | 2023-04-04 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYK
 
 | |
4ZNQ
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-2)Man | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | 登録日 | 2015-05-05 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
8IYL
 
 | |
4ZNO
 
 | | Crystal structure of Dln1 complexed with sucrose | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Natterin-like protein, ... | | 著者 | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | 登録日 | 2015-05-05 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNR
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-3)Man | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | 登録日 | 2015-05-05 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
8KGE
 
 | |
4TZ7
 
 | | Crystal structure of type I phosphatidylinositol 4-phosphate 5-kinase alpha from Zebrafish | | 分子名称: | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | | 著者 | Hu, J, Qin, Y, Wang, J, Li, L, Wu, D, Ha, Y. | | 登録日 | 2014-07-09 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled.
Nat Commun, 6, 2015
|
|
7R6Z
 
 | | OXA-48 bound by Compound 3.3 | | 分子名称: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | 登録日 | 2021-06-24 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
8JY6
 
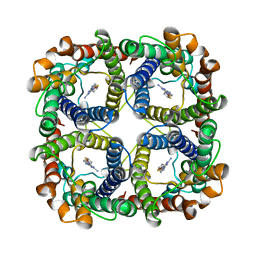 | | Structure of TbAQP2 in complex with anti-trypanosomatid drug melarsoprol | | 分子名称: | Aquaglyceroporin 2, [(4~{R})-2-[4-[[4,6-bis(azanyl)-1,3,5-triazin-2-yl]amino]phenyl]-1,3,2-dithiarsolan-4-yl]methanol | | 著者 | Chen, W, Wang, C. | | 登録日 | 2023-07-03 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural insights into drug transport by an aquaglyceroporin.
Nat Commun, 15, 2024
|
|
8JY7
 
 | |
8JY8
 
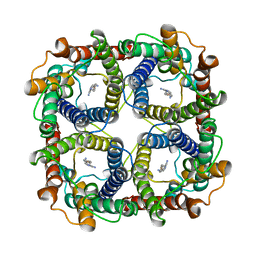 | |
8DZB
 
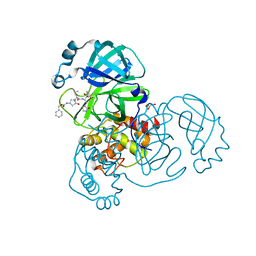 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 11 | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL, benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2022-08-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
8DZC
 
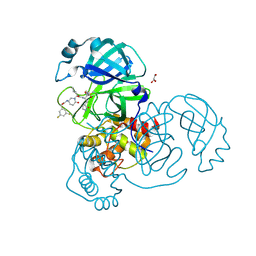 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 17 | | 分子名称: | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate, 3C-like proteinase nsp5, GLYCEROL | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2022-08-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
6EB0
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI | | 分子名称: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, ACETATE ION | | 著者 | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Yan, Y, Rose, J. | | 登録日 | 2018-08-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
