6TTV
 
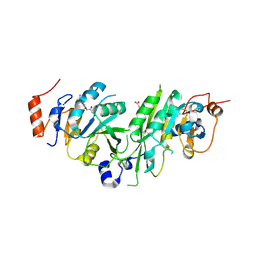 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 3 (ASI_M3M_138) | | 分子名称: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TUI
 
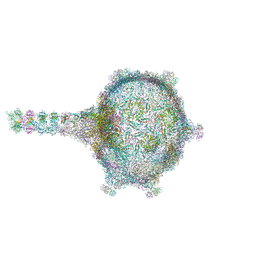 | | Virion of empty GTA particle | | 分子名称: | Adaptor protein Rcc01688, Phage major capsid protein, HK97 family, ... | | 著者 | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | 登録日 | 2020-01-07 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (10.47 Å) | | 主引用文献 | Virion of empty GTA particle
Nat Commun, 11, 2020
|
|
2O7M
 
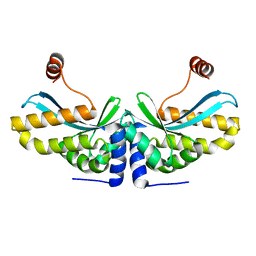 | | The C-terminal loop of the homing endonuclease I-CreI is essential for DNA binding and cleavage. Identification of a novel site for specificity engineering in the I-CreI scaffold | | 分子名称: | DNA endonuclease I-CreI | | 著者 | Prieto, J, Redondo, P, Padro, D, Blanco, F.J, Paques, F, Montoya, G. | | 登録日 | 2006-12-11 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The C-terminal loop of the homing endonuclease I-CreI is essential for site recognition, DNA binding and cleavage
Nucleic Acids Res., 35, 2007
|
|
1G0W
 
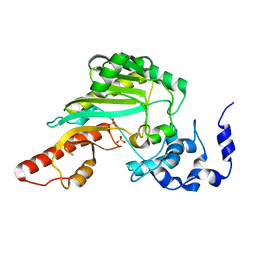 | |
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | 分子名称: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | 著者 | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-04 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
2O7S
 
 | |
3HOV
 
 | | Complete RNA polymerase II elongation complex II | | 分子名称: | 5'-D(*AP*GP*CP*TP*CP*AP*A*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*T*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', ... | | 著者 | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | 登録日 | 2009-06-03 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
3ER5
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | 分子名称: | ENDOTHIAPEPSIN, H-189 | | 著者 | Bailey, D, Veerapandian, B, Cooper, J, Szelke, M, Blundell, T.L. | | 登録日 | 1991-01-05 | | 公開日 | 1991-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Biochem.J., 289 ( Pt 2), 1993
|
|
2OJ2
 
 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | 分子名称: | Hematopoetic Cell Kinase, SH3 domain, artificial peptide PD1 | | 著者 | Schmidt, H, Hoffmann, S, Tran, T, Stoldt, M, Stangler, T, Wiesehan, K, Willbold, D. | | 登録日 | 2007-01-12 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a Hck SH3 Domain Ligand Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
4AEZ
 
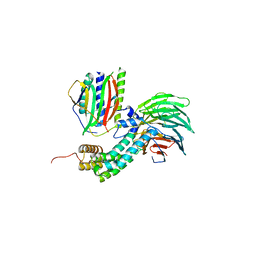 | | Crystal Structure of Mitotic Checkpoint Complex | | 分子名称: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | 著者 | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | 登録日 | 2012-01-13 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
6T4H
 
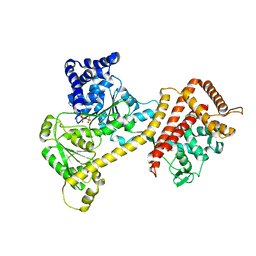 | | Crystal structure of the accessory translocation ATPase, SecA2, from Clostridium difficile, in complex with adenosine-5'-(gamma-thio)-triphosphate | | 分子名称: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein translocase subunit SecA 2 | | 著者 | Lindic, N, Loboda, J, Usenik, A, Turk, D. | | 登録日 | 2019-10-14 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Structure of Clostridioides difficile SecA2 ATPase Exposes Regions Responsible for Differential Target Recognition of the SecA1 and SecA2-Dependent Systems.
Int J Mol Sci, 21, 2020
|
|
3EII
 
 | | Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | 著者 | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | 登録日 | 2008-09-16 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3EJA
 
 | | Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | 登録日 | 2008-09-18 | | 公開日 | 2009-10-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
4AAU
 
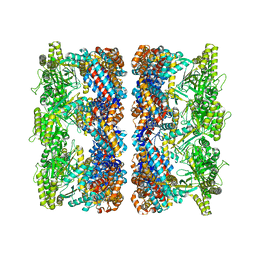 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | 分子名称: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | 登録日 | 2011-12-05 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
8BBG
 
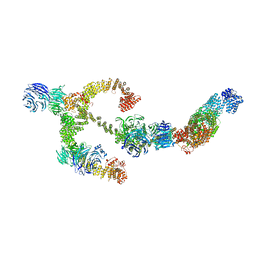 | | Structure of the IFT-A complex; anterograde IFT-A train model | | 分子名称: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | 著者 | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | 登録日 | 2022-10-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
6TFY
 
 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 18c | | 分子名称: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[5-[4-[[3-chloranyl-4-(pyridin-2-ylmethoxy)phenyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]-2-(3-oxidanylpropoxy)phenyl]propanamide | | 著者 | Niggenaber, J, Mueller, M.P, Rauh, D. | | 登録日 | 2019-11-14 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
3EKF
 
 | |
2NWX
 
 | | Crystal structure of GltPh in complex with L-aspartate and sodium ions | | 分子名称: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, PALMITIC ACID, ... | | 著者 | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | 登録日 | 2006-11-16 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
6THE
 
 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2019-11-20 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
4A2N
 
 | | Crystal Structure of Ma-ICMT | | 分子名称: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | 著者 | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | 登録日 | 2011-09-27 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
3E8V
 
 | | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis | | 分子名称: | Possible transglutaminase-family protein, UNKNOWN LIGAND | | 著者 | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-08-20 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis
To be Published
|
|
2O2F
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | 分子名称: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-2 | | 著者 | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | 登録日 | 2006-11-29 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
3HUV
 
 | |
4A2Y
 
 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH CITRATE ANIONS | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN | | 著者 | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | 登録日 | 2011-09-29 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
1FVT
 
 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | 分子名称: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | 登録日 | 2000-09-20 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
