7DPF
 
 | |
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
5UKO
 
 | |
8OYP
 
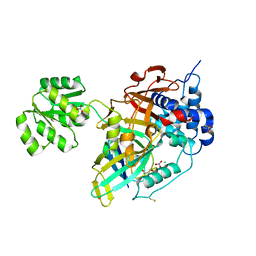 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | 分子名称: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | 登録日 | 2023-05-05 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
5UKQ
 
 | |
7Y3G
 
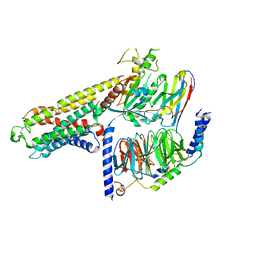 | | Cryo-EM structure of a class A orphan GPCR | | 分子名称: | G-protein coupled receptor 12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, Z.J, Hua, T, Li, H, Zhang, J.Y, Luo, F. | | 登録日 | 2022-06-10 | | 公開日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural insight into the constitutive activity of human orphan receptor GPR12.
Sci Bull (Beijing), 68, 2023
|
|
5UDE
 
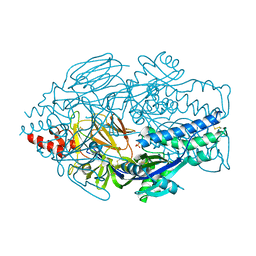 | | Crystal Structure of RSV F B9320 DS-Cav1 | | 分子名称: | Fusion glycoprotein F0, SULFATE ION | | 著者 | McLellan, J.S. | | 登録日 | 2016-12-26 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
5UKP
 
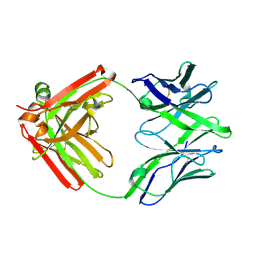 | |
5UKN
 
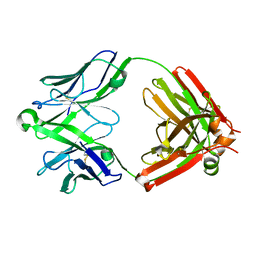 | |
7X78
 
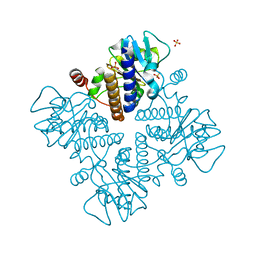 | | L-fuculose 1-phosphate aldolase | | 分子名称: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | 著者 | Lou, X, Zhang, Q, Bartlam, M. | | 登録日 | 2022-03-09 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
6ISO
 
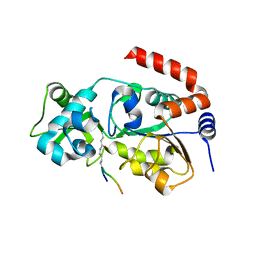 | | Human SIRT3 Recognizing H3K4cr | | 分子名称: | (2E)-BUT-2-ENAL, ARG-THR-LYS-GLN-THR-ALA-ARG, GLYCEROL, ... | | 著者 | Wang, Y, Hao, Q. | | 登録日 | 2018-11-17 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Elife, 3, 2014
|
|
8J0K
 
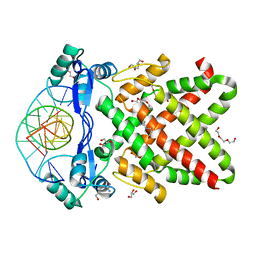 | | Crystal structure of human TFAP2A in complex with DNA | | 分子名称: | DNA (5'-D(*CP*TP*GP*CP*CP*TP*CP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*CP*GP*AP*GP*GP*CP*AP*G)-3'), GLYCEROL, ... | | 著者 | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | 登録日 | 2023-04-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0Q
 
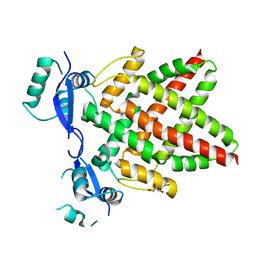 | | Structure of DNA binding domain of human TFAP2B | | 分子名称: | GLYCEROL, Transcription factor AP-2-beta | | 著者 | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | 登録日 | 2023-04-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0L
 
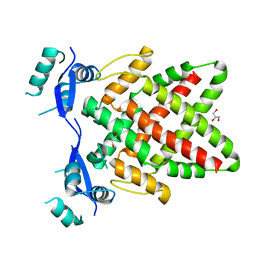 | | Structure of DNA binding Domain of Human TFAP2A | | 分子名称: | GLYCEROL, Transcription factor AP-2-alpha | | 著者 | Liu, K, Xiao, Y.Q, Gan, L.Y, Min, J.R. | | 登録日 | 2023-04-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0R
 
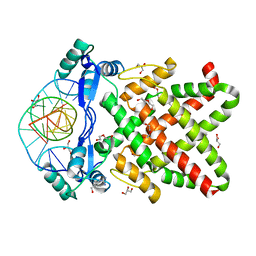 | | Structure of human TFAP2A in complex with DNA | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*CP*CP*TP*CP*AP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*TP*GP*AP*GP*GP*CP*AP*G)-3'), ... | | 著者 | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | 登録日 | 2023-04-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
2L3T
 
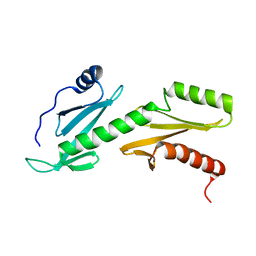 | |
8GKA
 
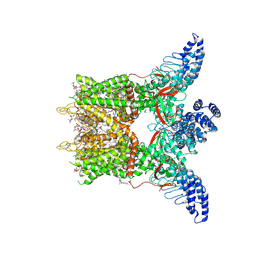 | | Human TRPV3 tetramer structure, closed conformation | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | 著者 | Lansky, S, Betancourt, J.M, Scheuring, S. | | 登録日 | 2023-03-17 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | A pentameric TRPV3 channel with a dilated pore.
Nature, 621, 2023
|
|
8GKG
 
 | |
8HAW
 
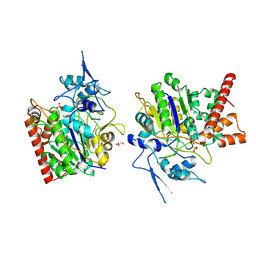 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8H3X
 
 | |
8H3Y
 
 | |
