7UO0
 
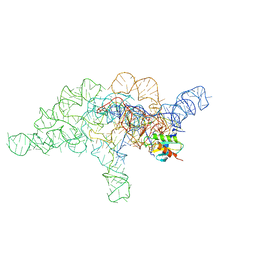 | | E.coli RNaseP Holoenzyme with Mg2+ | | 分子名称: | CALCIUM ION, Precursor tRNA substrate G(-1) G(-2), RNase P RNA, ... | | 著者 | Huang, W, Taylor, D.J. | | 登録日 | 2022-04-12 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO1
 
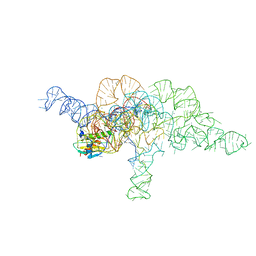 | | E.coli RNaseP Holoenzyme with Mg2+ | | 分子名称: | CALCIUM ION, E.coli RNase P RNA, Precursor tRNA substrate U(-1) and A(-2), ... | | 著者 | Huang, W, Taylor, D.J. | | 登録日 | 2022-04-12 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
5K8K
 
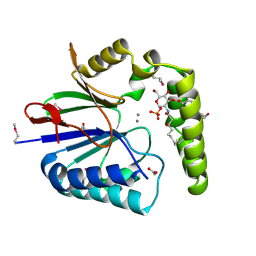 | | Structure of the Haemophilus influenzae LpxH-lipid X complex | | 分子名称: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, ACETATE ION, GLYCEROL, ... | | 著者 | Cho, J, Lee, C.-J, Zhou, P. | | 登録日 | 2016-05-30 | | 公開日 | 2016-08-10 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Nat Microbiol, 1, 2016
|
|
7UO2
 
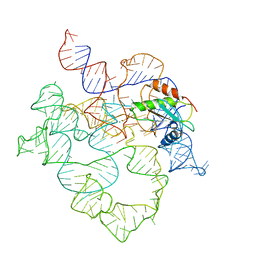 | | E.coli RNaseP Holoenzyme with Mg2+ | | 分子名称: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component | | 著者 | Huang, W, Taylor, D.J. | | 登録日 | 2022-04-12 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO5
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | 分子名称: | CALCIUM ION, RNase P RNA, Ribonuclease P protein component | | 著者 | Huang, W, Taylor, D.J. | | 登録日 | 2022-04-12 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
8SBG
 
 | |
8SIJ
 
 | |
8SL7
 
 | |
6PSK
 
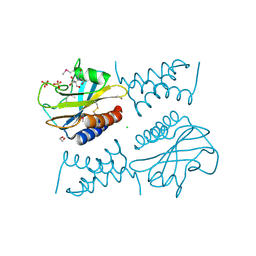 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | 著者 | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | 登録日 | 2019-07-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2020-08-12 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
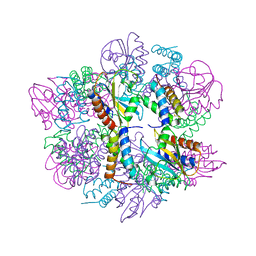
6PX4
 
 | |
8TQD
 
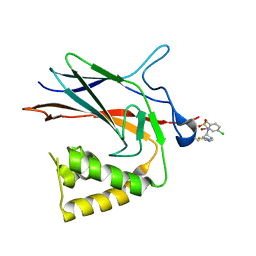 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | 分子名称: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | 著者 | Hilbert, B.J. | | 登録日 | 2023-08-07 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
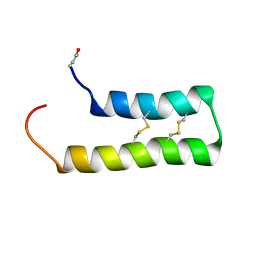
6WQL
 
 | |
6WQJ
 
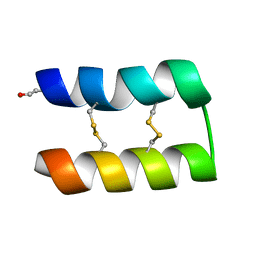 | |
8T5K
 
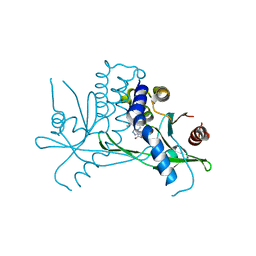 | | Crystal structure of STING CTD in complex with BDW-OH | | 分子名称: | Stimulator of interferon genes protein, {[(4S)-8,9-dimethylthieno[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-3-yl]sulfanyl}acetic acid | | 著者 | Li, Y, Li, P, Sun, D. | | 登録日 | 2023-06-13 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8T5L
 
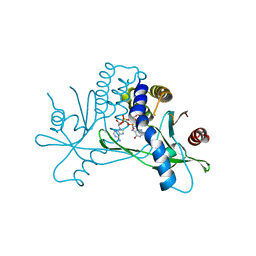 | |
6WC5
 
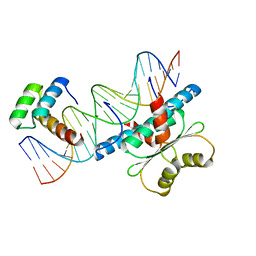 | |
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
3MTS
 
 | | Chromo Domain of Human Histone-Lysine N-Methyltransferase SUV39H1 | | 分子名称: | Histone-lysine N-methyltransferase SUV39H1 | | 著者 | Lam, R, Li, Z, Wang, J, Crombet, L, Walker, J.R, Ouyang, H, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-04-30 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Human SUV39H1 Chromodomain and Its Recognition of Histone H3K9me2/3.
Plos One, 7, 2012
|
|
3IGV
 
 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | 分子名称: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | 著者 | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | 登録日 | 2009-07-28 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4QUW
 
 | |
7MJR
 
 | | Vip4Da2 toxin structure | | 分子名称: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | 著者 | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
4RC7
 
 | |
4RC8
 
 | |
7JWL
 
 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | 分子名称: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | 登録日 | 2020-08-25 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
4RC5
 
 | |
