8R8Q
 
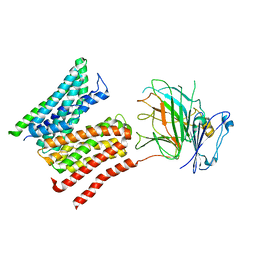 | | Lysosomal peptide transporter | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosylated lysosomal membrane protein, Major facilitator superfamily domain-containing protein 1 | | 著者 | Jungnickel, K.E.J, Loew, C. | | 登録日 | 2023-11-29 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.08 Å) | | 主引用文献 | MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes
Nature Cell Biology, 2024
|
|
8K5Y
 
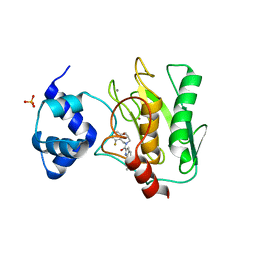 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
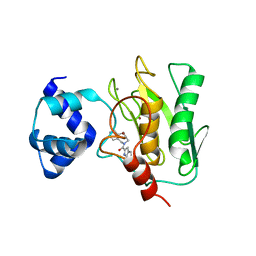 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5W
 
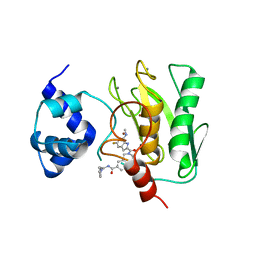 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
1TCG
 
 | |
1TCJ
 
 | |
1HJB
 
 | |
1HJC
 
 | |
1IO4
 
 | |
1TCK
 
 | |
1TCH
 
 | |
8K4Z
 
 | |
8A51
 
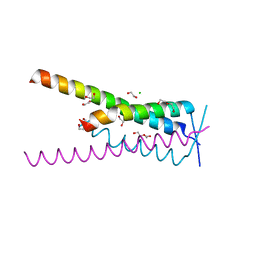 | | Crystal structure of HSF2BP-BRME1 complex | | 分子名称: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | 著者 | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | 登録日 | 2022-06-13 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
8A50
 
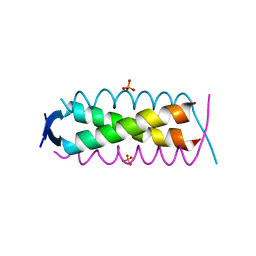 | | Crystal structure of HSF2BP-ALPHA1 tetramer | | 分子名称: | Heat shock factor 2-binding protein, PHOSPHATE ION | | 著者 | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | 登録日 | 2022-06-13 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.484 Å) | | 主引用文献 | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
1HSR
 
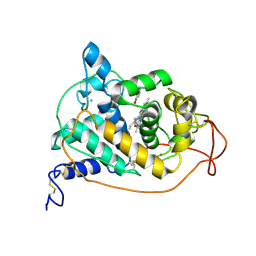 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | 著者 | Fukuyama, K, Itakura, H. | | 登録日 | 1997-07-01 | | 公開日 | 1998-07-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Suzuki, T, Kimura, K, Hashiguchi, T. | | 登録日 | 2022-05-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
5B5P
 
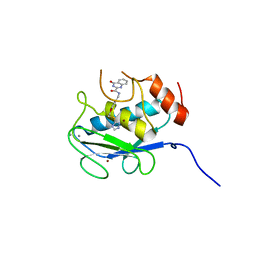 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-oxo-N-(3-(2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy)benzyl)-3,4-dihydroquinazoline-2-carboxamide | | 分子名称: | 4-oxo-N-{3-[2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy]benzyl}-3,4-dihydroquinazoline-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | 著者 | Oki, H, Tanaka, Y. | | 登録日 | 2016-05-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
1GVD
 
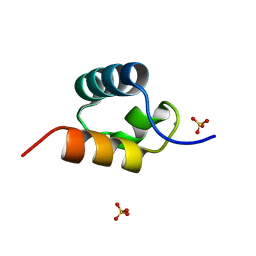 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | 分子名称: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-02-08 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV5
 
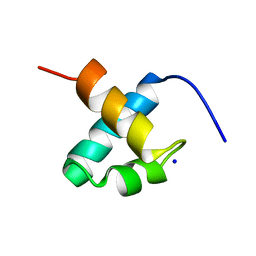 | | CRYSTAL STRUCTURE OF C-MYB R2 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-02-06 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
5GXQ
 
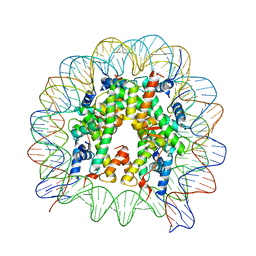 | | The crystal structure of the nucleosome containing H3.6 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2016-09-19 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
1GUU
 
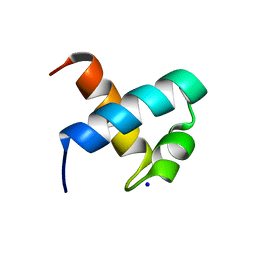 | | CRYSTAL STRUCTURE OF C-MYB R1 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-01-30 | | 公開日 | 2003-06-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-02-05 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
