6LVP
 
 | |
6LVO
 
 | |
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 2004-05-30 | | 公開日 | 2005-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W01
 
 | |
1W02
 
 | |
7JTH
 
 | |
7K01
 
 | | Structure of TFIIH in TFIIH/Rad4-Rad23-Rad33 DNA opening complex | | 分子名称: | DNA repair helicase RAD25, DNA repair helicase RAD3, General transcription and DNA repair factor IIH subunit SSL1, ... | | 著者 | van Eeuwen, T, Min, J.H, Murakami, K. | | 登録日 | 2020-09-02 | | 公開日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7K04
 
 | | Structure of TFIIH/Rad4-Rad23-Rad33/DNA in DNA opening | | 分子名称: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | 著者 | van Eeuwen, T, Min, J.H, Murakami, K. | | 登録日 | 2020-09-03 | | 公開日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (9.25 Å) | | 主引用文献 | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7KUE
 
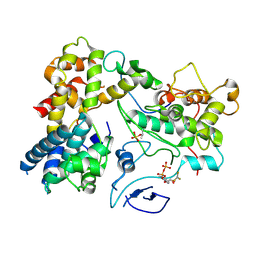 | | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | 著者 | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | 登録日 | 2020-11-24 | | 公開日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
5Y7K
 
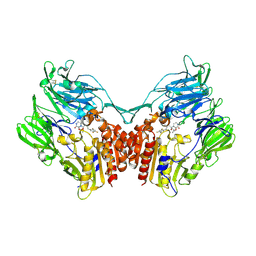 | | Crystal structure of human DPP4 in complex with inhibitor1 | | 分子名称: | (R)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazine-2-one, Dipeptidyl peptidase 4 | | 著者 | Lee, H.K, Kim, E.E. | | 登録日 | 2017-08-17 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.512 Å) | | 主引用文献 | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7J
 
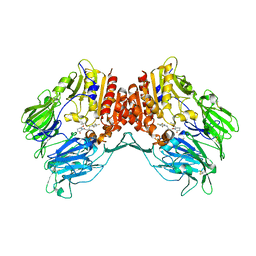 | | Crystal structure of human DPP4 in complex with inhibitor2 | | 分子名称: | (S)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazin-2-one, Dipeptidyl peptidase 4 | | 著者 | Lee, H.K, Kim, E.E. | | 登録日 | 2017-08-17 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.521 Å) | | 主引用文献 | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7H
 
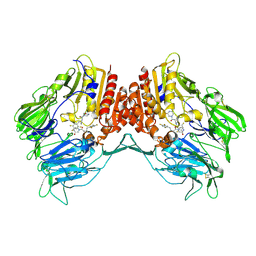 | |
5XV9
 
 | |
5Y86
 
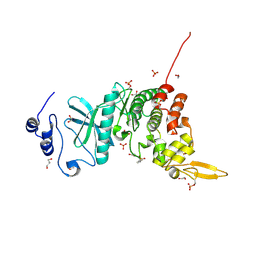 | | Crystal structure of kinase | | 分子名称: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | 著者 | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | 登録日 | 2017-08-18 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
8TH1
 
 | |
8TH7
 
 | |
8TH6
 
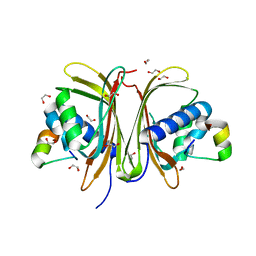 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | 分子名称: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | 著者 | Hughes, M.P, Taylor, J.P, Yang, Z. | | 登録日 | 2023-07-14 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
8TH5
 
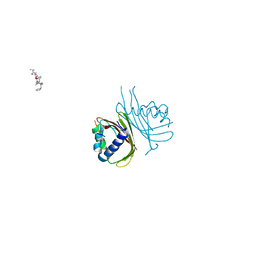 | |
8SGH
 
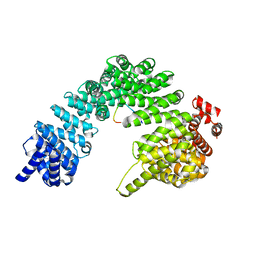 | |
2MJ6
 
 | |
8DLF
 
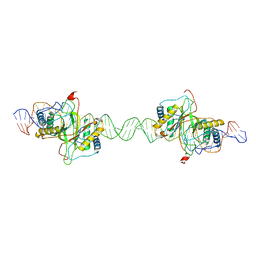 | |
5XIX
 
 | |
6INT
 
 | | xylose isomerase from Paenibacillus sp. R4 | | 分子名称: | CALCIUM ION, Xylose isomerase | | 著者 | Lee, J.H, Lee, C.W, Park, S. | | 登録日 | 2018-10-26 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Xylose Isomerase (PbXI) from the Psychrophilic Soil Microorganism, Paenibacillus sp.
J. Microbiol. Biotechnol., 29, 2019
|
|
2POO
 
 | |
1SMA
 
 | |
