2I5T
 
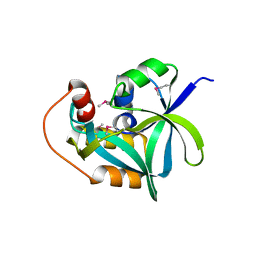 | | Crystal Structure of hypothetical protein LOC79017 from Homo sapiens | | 分子名称: | Protein C7orf24 | | 著者 | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-08-25 | | 公開日 | 2006-09-12 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
2I3C
 
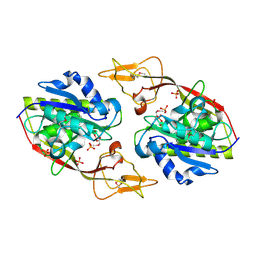 | | Crystal Structure of an Aspartoacylase from Homo Sapiens | | 分子名称: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | 著者 | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-08-17 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I5S
 
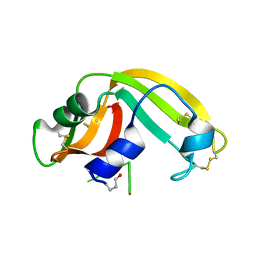 | | Crystal structure of onconase with bound nucleic acid | | 分子名称: | 5'-D(*A*(DU)P*GP*A)-3', P-30 protein | | 著者 | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-08-25 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
2GNX
 
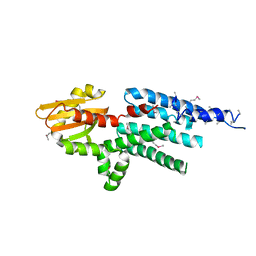 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | 分子名称: | hypothetical protein | | 著者 | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-11 | | 公開日 | 2006-05-02 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
2H1S
 
 | |
2GMK
 
 | | Crystal structure of onconase double mutant with spontaneously-assembled (AMP) 4 stack | | 分子名称: | ADENOSINE MONOPHOSPHATE, P-30 protein | | 著者 | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-06 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
2GM3
 
 | | Crystal Structure of an Universal Stress Protein Family Protein from Arabidopsis Thaliana At3g01520 with AMP Bound | | 分子名称: | ADENOSINE MONOPHOSPHATE, unknown protein | | 著者 | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-05 | | 公開日 | 2006-04-18 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.461 Å) | | 主引用文献 | Crystal structure of the protein At3g01520, a eukaryotic universal stress protein-like protein from arabidopsis thaliana in complex with AMP.
Proteins, 83, 2015
|
|
2HO4
 
 | | Crystal Structure of Protein from Mouse Mm.236127 | | 分子名称: | Haloacid dehalogenase-like hydrolase domain containing 2, MAGNESIUM ION, PHOSPHATE ION | | 著者 | McCoy, J.G, Wesenberg, G.E, Bitto, E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-07-13 | | 公開日 | 2006-08-15 | | 最終更新日 | 2018-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Protein from Mouse Mm.236127
To be published
|
|
2I2O
 
 | | Crystal Structure of an eIF4G-like Protein from Danio rerio | | 分子名称: | NICKEL (II) ION, eIF4G-like protein | | 著者 | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-08-16 | | 公開日 | 2006-08-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure of an eIF4G-like protein from Danio rerio.
Proteins, 78, 2010
|
|
2GU2
 
 | | Crystal Structure of an Aspartoacylase from Rattus norvegicus | | 分子名称: | Aspa protein, SULFATE ION, ZINC ION | | 著者 | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-28 | | 公開日 | 2006-06-20 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HB5
 
 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | 分子名称: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | 著者 | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | 登録日 | 2006-06-13 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
4KX5
 
 | |
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | 著者 | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-08 | | 公開日 | 2013-09-04 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4L2Z
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | 分子名称: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-06-05 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | 分子名称: | Oleandomycin glycosyltransferase, SODIUM ION | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4O89
 
 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | 分子名称: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | 著者 | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | 登録日 | 2013-12-26 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
4O8J
 
 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii, in complex with rACAAA3'phosphate and adenine. | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, RNA, ... | | 著者 | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | 登録日 | 2013-12-27 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | 分子名称: | uncharacterized protein SgcJ | | 著者 | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-11-20 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.719 Å) | | 主引用文献 | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | 分子名称: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | 著者 | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-01-29 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 | | 著者 | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4N2P
 
 | | Structure of Archease from Pyrococcus horikoshii | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | 登録日 | 2013-10-05 | | 公開日 | 2014-01-01 | | 最終更新日 | 2020-09-23 | | 実験手法 | X-RAY DIFFRACTION (1.435 Å) | | 主引用文献 | A tRNA splicing operon: Archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation.
Nucleic Acids Res., 42, 2014
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | 分子名称: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4PIW
 
 | | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12 | | 分子名称: | TDP-4-keto-6-deoxy-D-glucose transaminase family protein | | 著者 | Wang, F, Xu, W, Helmich, K.E, Singh, S, Yennamalli, R.M, Miller, M.D, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-05-09 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12
To Be Published
|
|
4HPV
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus | | 分子名称: | S-adenosylmethionine synthase | | 著者 | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-10-24 | | 公開日 | 2012-11-14 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.214 Å) | | 主引用文献 | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | 分子名称: | SULFATE ION, TlmII | | 著者 | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-06 | | 公開日 | 2012-11-21 | | 最終更新日 | 2014-04-16 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
