8JQ5
 
 | |
8JQ4
 
 | |
6T01
 
 | | Crystal structure of YTHDC1 with fragment 13 (DHU_DC1_153) | | 分子名称: | 6-methyl-5-nitro-4-phenyl-1~{H}-pyrimidin-2-one, SULFATE ION, YTH domain-containing protein 1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0A
 
 | | Crystal structure of YTHDC1 with fragment 25 (PSI_DC1_005) | | 分子名称: | (~{R})-azanyl(pyridin-3-yl)methanethiol, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
8JQ6
 
 | |
8JQ3
 
 | |
6T13
 
 | | CRYSTAL STRUCTURE OF GLUCOCEREBROSIDASE IN COMPLEX WITH A PYRROLOPYRAZINE | | 分子名称: | 1,2-ETHANEDIOL, 1-[4-[2-(4-methoxyphenyl)-5-methyl-pyrrolo[2,3-b]pyrazin-6-yl]piperidin-1-yl]ethanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Benz, J, Ehler, A, Hug, M, Huber, S, Rufer, A.C, Guba, W, Jagasia, R, Hofmann, E.C, Rodriguez Sarmiento, R.M. | | 登録日 | 2019-10-03 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Novel beta-Glucocerebrosidase Activators That Bind to a New Pocket at a Dimer Interface and Induce Dimerization.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y1V
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide | | 分子名称: | (2R)-2-[(2,6-diethylphenyl)carbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide
To Be Published
|
|
8J4C
 
 | | YeeE(TsuA)-YeeD(TsuB) complex for thiosulfate uptake | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Spirochaeta thermophila YeeE(TsuA)-YeeD(TsuB),UPF0033 domain-containing protein, SirA-like domain-containing protein (chimera), ... | | 著者 | Ikei, M, Miyazaki, R, Monden, K, Naito, Y, Takeuchi, A, Takahashi, Y.S, Tanaka, Y, Ichikawa, M, Tsukazaki, T. | | 登録日 | 2023-04-19 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Structure and function of YeeE-YeeD complex for sophisticated thiosulfate uptake
To Be Published
|
|
2BZG
 
 | | Crystal structure of thiopurine S-methyltransferase. | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | 著者 | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | 登録日 | 2005-08-17 | | 公開日 | 2005-08-25 | | 最終更新日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
1RXF
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH FE(II) | | 分子名称: | DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | 著者 | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | 登録日 | 1998-06-05 | | 公開日 | 1999-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
6SZY
 
 | | Crystal structure of YTHDC1 with fragment 12 (DHU_DC1_150) | | 分子名称: | 4-propan-2-yl-5,6,7,8-tetrahydro-4~{a}~{H}-quinazolin-2-one, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T04
 
 | | Crystal structure of YTHDC1 with fragment 17 (DHU_DC1_042) | | 分子名称: | 2-phenyl-4,5-dihydro-1~{H}-imidazole, SULFATE ION, YTH domain-containing protein 1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5YO1
 
 | | Structure of ePepN E298A mutant in complex with Puromycin | | 分子名称: | Aminopeptidase N, GLYCEROL, PUROMYCIN, ... | | 著者 | Ganji, R.J, Reddi, R, Marapaka, A.K, Addlagatta, A. | | 登録日 | 2017-10-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Puromycin, a selective inhibitor of PSA acts as a substrate for other M1 family aminopeptidases: Biochemical and structural basis
Int.J.Biol.Macromol., 165, 2020
|
|
5YP1
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
4E70
 
 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol | | 分子名称: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, GLYCEROL | | 著者 | Wolters, S, Heine, A, Petersen, M. | | 登録日 | 2012-03-16 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6093 Å) | | 主引用文献 | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6SZ2
 
 | | Crystal structure of YTHDC1 with fragment 3 (DHU_DC1_149) | | 分子名称: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methyl-1,6-naphthyridin-4-amine | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-01 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4Y28
 
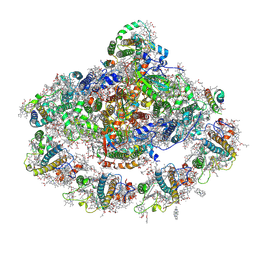 | | The structure of plant photosystem I super-complex at 2.8 angstrom resolution. | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Mazor, Y, Brovikov, A, Nelson, N. | | 登録日 | 2015-02-09 | | 公開日 | 2015-08-19 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structure of plant photosystem I super-complex at 2.8 angstrom resolution.
Elife, 4, 2015
|
|
6SZ9
 
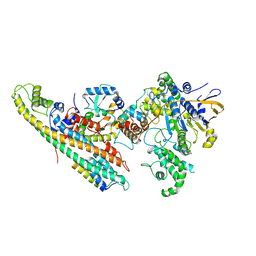 | | Type IV Coupling Complex (T4CC) from L. pneumophila. | | 分子名称: | DotY, DotZ, IcmJ (DotN), ... | | 著者 | Mace, K, Meir, A, Lukoyanova, N, Waksman, G. | | 登録日 | 2019-10-02 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Mechanism of effector capture and delivery by the type IV secretion system from Legionella pneumophila.
Nat Commun, 11, 2020
|
|
5YR6
 
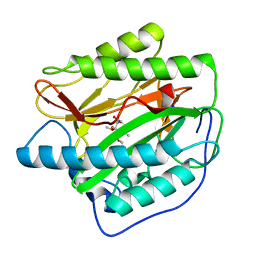 | | Human methionine aminopeptidase type 1b (F309L mutant) in complex with TNP470 | | 分子名称: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, Methionine aminopeptidase 1, ... | | 著者 | Pillalamarri, V, Arya, T, Addlagatta, A. | | 登録日 | 2017-11-08 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of natural product ovalicin sensitive type 1 methionine aminopeptidases: molecular and structural basis.
Biochem. J., 476, 2019
|
|
8JOR
 
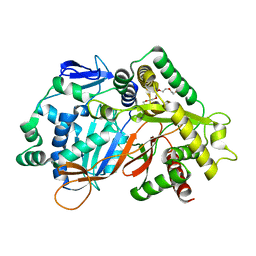 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | 分子名称: | Acyltransferase, PENTAETHYLENE GLYCOL | | 著者 | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | 登録日 | 2023-06-08 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
5YAQ
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with scyllo-inosose | | 分子名称: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | 著者 | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2017-09-01 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
1S9U
 
 | | Atomic structure of a putative anaerobic dehydrogenase component | | 分子名称: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | 著者 | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-02-05 | | 公開日 | 2004-06-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
8JOS
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | 分子名称: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | 著者 | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | 登録日 | 2023-06-08 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
